The hidden duplication past of Arabidopsis thaliana
- PMID: 12374856
- PMCID: PMC129725
- DOI: 10.1073/pnas.212522399
The hidden duplication past of Arabidopsis thaliana
Abstract
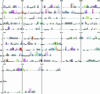
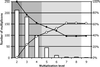
Analysis of the genome sequence of Arabidopsis thaliana shows that this genome, like that of many other eukaryotic organisms, has undergone large-scale gene duplications or even duplications of the entire genome. However, the high frequency of gene loss after duplication events reduces colinearity and therefore the chance of finding duplicated regions that, at the extreme, no longer share homologous genes. In this study we show that heavily degenerated block duplications that can no longer be recognized by directly comparing two segments because of differential gene loss, can still be detected through indirect comparison with other segments. When these so-called hidden duplications in Arabidopsis are taken into account, many homologous genomic regions can be found in five to eight copies. This finding strongly implies that Arabidopsis has undergone three, but probably no more, rounds of genome duplications. Therefore, adding such hidden blocks to the duplication landscape of Arabidopsis sheds light on the number of polyploidy events that this model plant genome has undergone in its evolutionary past.
Figures


References
-
- Terryn N., Heijnen, L., De Keyser, A., Van Asseldonck, M., De Clercq, R., Verbakel, H., Gielen, J., Zabeau, M., Villarroel, R., Jesse, T., et al. (1999) FEBS Lett. 445, 237-245. - PubMed
-
- The Arabidopsis Genome Initiative (2000) Nature 408, 796-815. - PubMed
-
- Lynch M. & Conery, J. S. (2000) Science 290, 1151-1155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

