Cell surface polarization during yeast mating
- PMID: 12374868
- PMCID: PMC137858
- DOI: 10.1073/pnas.172517799
Cell surface polarization during yeast mating
Abstract
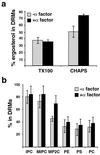
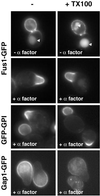
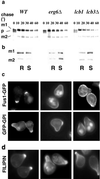
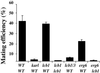
Exposure to mating pheromone in haploid Saccharomyces cerevisiae cells results in the arrest of the cell cycle, expression of mating-specific genes, and polarized growth toward the mating partner. Proteins involved in signaling, polarization, cell adhesion, and fusion are localized to the tip of the mating cell (shmoo) where fusion will eventually occur. The mechanisms ensuring the correct targeting and retention of these proteins are poorly understood. Here we show that in pheromone-treated cells, a reorganization of the plasma membrane involving lipid rafts results in the retention of proteins at the tip of the mating projection, segregated from the rest of the membrane. Sphingolipid and ergosterol biosynthetic mutants fail to polarize proteins to the tip of the shmoo and are deficient in mating. Our results show that membrane microdomain clustering at the mating projection is involved in the generation and maintenance of polarity during mating.
Figures



References
-
- van Meer G., Gumbiner, B. & Simons, K. (1986) Nature 322, 639-641. - PubMed
-
- Bromley S. K., Burack, W. R., Johnson, K. G., Somersalo, K., Sims, T. N., Sumen, C., Davis, M. M., Shaw, A. S., Allen, P. M. & Dustin, M. L. (2001) Annu. Rev. Immunol. 19, 375-396. - PubMed
-
- Brown D. A. & London, E. (1998) Annu. Rev. Cell Dev. Biol. 14, 111-136. - PubMed
-
- Simons K. & Toomre, D. (2000) Nat. Rev. Mol. Cell. Biol. 1, 31-39. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

