Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula
- PMID: 12376622
- PMCID: PMC166584
- DOI: 10.1104/pp.006833
Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula
Abstract
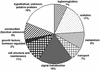
The Medicago truncatula expressed sequence tag (EST) database (Gene Index) contains over 140,000 sequences from 30 cDNA libraries. This resource offers the possibility of identifying previously uncharacterized genes and assessing the frequency and tissue specificity of their expression in silico. Because M. truncatula forms symbiotic root nodules, unlike Arabidopsis, this is a particularly important approach in investigating genes specific to nodule development and function in legumes. Our analyses have revealed 340 putative gene products, or tentative consensus sequences (TCs), expressed solely in root nodules. These TCs were represented by two to 379 ESTs. Of these TCs, 3% appear to encode novel proteins, 57% encode proteins with a weak similarity to the GenBank accessions, and 40% encode proteins with strong similarity to the known proteins. Nodule-specific TCs were grouped into nine categories based on the predicted function of their protein products. Besides previously characterized nodulins, other examples of highly abundant nodule-specific transcripts include plantacyanin, agglutinin, embryo-specific protein, and purine permease. Six nodule-specific TCs encode calmodulin-like proteins that possess a unique cleavable transit sequence potentially targeting the protein into the peribacteroid space. Surprisingly, 114 nodule-specific TCs encode small Cys cluster proteins with a cleavable transit peptide. To determine the validity of the in silico analysis, expression of 91 putative nodule-specific TCs was analyzed by macroarray and RNA-blot hybridizations. Nodule-enhanced expression was confirmed experimentally for the TCs composed of five or more ESTs, whereas the results for those TCs containing fewer ESTs were variable.
Figures



References
-
- Aalen RB, Opsahlferstad HG, Linnestad C, Olsen OA. Transcripts encoding an oleosin and a dormancy-related protein are present in both the aleurone layer and the embryo of developing barley (Hordeum vulgareL.) seeds. Plant J. 1994;5:385–396. - PubMed
-
- Ablett E, Seaton G, Scott K, Shelton D, Graham MW, Baverstock P, SladeLee L, Henry R. Analysis of grape ESTs: global gene expression patterns in leaf and berry. Plant Sci. 2000;159:87–95. - PubMed
-
- AGI. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

