Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis
- PMID: 12376625
- PMCID: PMC166587
- DOI: 10.1104/pp.006189
Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis
Abstract
Proanthocyanidin (PA), or condensed tannin, is a polymeric flavanol that accumulates in a number of tissues in a wide variety of plants. In Arabidopsis, we found that PA precursors (detected histochemically using OsO(4)) accumulate in the endothelial cell layer of the seed coat from the two-terminal cell stage of embryo development onwards. To understand how PA is made, we screened mature seed pools of T-DNA-tagged Arabidopsis lines to identify mutants defective in the synthesis of PA and found six tds (tannin-deficient seed) complementation groups defective in PA synthesis. Mutations in these loci disrupt the amount (tds1, tds2, tds3, tds5, and tds6) or location and amount of PA (tds4) in the endothelial cell layer. The PA intermediate epicatechin has been identified in wild type and mutants tds1, tds2, tds3, and tds5 (which do not produce PA) and tds6 (6% of wild-type PA), whereas tds4 (2% of wild-type PA) produces an unidentified dimethylaminocinnamaldehyde-reacting compound, indicating that the mutations may be acting on genes beyond leucoanthocyanidin reductase, the first enzymatic reduction step dedicated to PA synthesis. Two other mutants were identified, an allele of tt7, which has a spotted pattern of PA deposition and produces only 8% of the wild-type level of type PA as propelargonidin, and an allele of tt8 producing no PA. Spotted patterns of PA deposition observed in seed of mutants tds4 and tt7-3 result from altered PA composition and distribution in the cell. Our mutant screen, which was not exhaustive, suggests that the cooperation of many genes is required for successful PA accumulation.
Figures
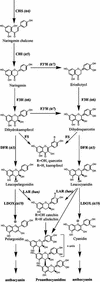





References
-
- Albert S, Delseny M, Devic M. BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J. 1997;11:289–299. - PubMed
-
- Andersen OM, Francis GW. Simultaneous analysis of anthocyanins and anthocyanidins on cellulose thin layers. J Chromatogr. 1985;318:450–454.
-
- Baur PS, Walkinshaw CH. Fine structure of tannin accumulations in callus cultures of Pinus elliotti (slash pine) Can J Bot. 1974;52:615–619.
-
- Bethke PC, Jones RL. Vacuoles and prevacuolar compartments. Curr Opin Plant Biol. 2000;3:469–475. - PubMed
-
- Chafe SC, Durzan DJ. Tannin inclusions in cell suspension cultures of white spruce. Planta. 1973;113:251–262. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

