Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction
- PMID: 12376626
- PMCID: PMC166588
- DOI: 10.1104/pp.009175
Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction
Abstract
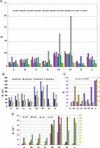
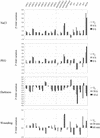
The recent publication of the complete sequence of the Arabidopsis genome allowed us to identify and characterize the last two members of the SHAGGY-like kinase (AtSK) gene family. As a result, the study of the overall spatio-temporal organization of the whole AtSK family in Arabidopsis has become an achievable and necessary aim to understand the role of each SHAGGY-like kinase during plant development. An analysis of the transcript level of the 10 members of the family has been performed using the technique of real-time quantitative reverse transcriptase-polymerase chain reaction. Transcript levels in several organs, under different growth conditions, were analyzed. To calibrate the results obtained, a number of other genes, such as those coding for the two MAP3Kepsilons and the two MAP4Kalphas, as well as the stress response marker RD29A; the small subunit of the Rubisco photosynthetic enzyme Ats1A; the MEDEA chromatin remodeling factor; and the SCARECROW, ASYMMETRIC LEAVES 1, and SUPERMAN transcription factors all involved in key steps of plant development were used. The analysis of our data revealed that eight of the 10 genes of the AtSK family displayed a pseudo-constitutive expression pattern at the organ level. Conversely, AtSK13 responded to osmotic changes and saline treatment, whereas AtSK31 was flower specific and responded to osmotic changes and darkness.
Figures



References
-
- Amador V, Monte E, Garcia-Martinez JL, Prat S. Gibberellins signal nuclear import of PHOR1 a photoperiod-responsive protein with homology to Drosophilaarmadillo. Cell. 2001;106:343–354. - PubMed
-
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8actin subclass in vegetative tissues. Plant J. 1996;10:107–121. - PubMed
-
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. - PubMed
-
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. ASYMMETRIC LEAVES 1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. - PubMed
-
- Chikano H, Ogawa M, Ikeda Y, Koizumi N, Kusano T, Sano H. Two novel genes encoding SNF-1 related protein kinases from Arabidopsis thaliana: differential accumulation of AtSR1 and AtSR2transcripts in response to cytokinins and sugars and phosphorylation of sucrose synthase by AtSR2. Mol Gen Genet. 2001;264:674–681. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

