Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities
- PMID: 12376636
- PMCID: PMC166598
- DOI: 10.1104/pp.003566
Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities
Abstract
A small family of novel basic leucine zipper proteins that includes abscisic acid (ABA)-INSENSITIVE 5 (ABI5) binds to the promoter region of the lea class gene Dc3. The factors, referred to as AtDPBFs (Arabidopsis Dc3 promoter-binding factors), were isolated from an immature seed cDNA library. AtDPBFs bind to the embryo specification and ABA-responsive elements in the Dc3 promoter and are unique in that they can interact with cis-elements that do not contain the ACGT core sequence required for the binding of most other plant basic leucine zipper proteins. Analysis of full-length cDNAs showed that at least five different Dc3 promoter-binding factors are present in Arabidopsis seeds; one of these, AtDPBF-1, is identical to ABI5. As expected, AtDPBF-1/ABI5 mRNA is inducible by exogenous ABA in seedlings. Despite the near identity in their basic domains, AtDPBFs are distinct in their DNA-binding, dimerization, and transcriptional activity.
Figures


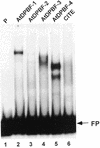
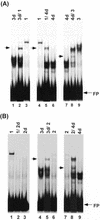

References
-
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: Green Publishing Associates/Wiley Interscience; 1994.
-
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci. 1993;316:1194–1199.
-
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

