ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis
- PMID: 12376638
- PMCID: PMC166600
- DOI: 10.1104/pp.009993
ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis
Abstract
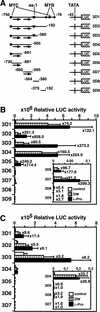
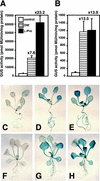
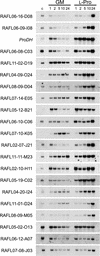
Proline (Pro) is one of the most widely distributed osmolytes in water-stressed plants. We previously isolated from Arabidopsis a gene encoding Pro dehydrogenase (ProDH), a mitochondrial enzyme involved in the first step of the conversion of Pro to glutamic acid. The ProDH gene in Arabidopsis is up-regulated by rehydration after dehydration but is down-regulated by dehydration. ProDH is also induced by L-Pro and hypoosmolarity. The induction of ProDH expression under rehydration seems to be caused by both accumulated Pro and hypoosmolarity. We analyzed a DNA region that is located 5' to the transcription start site (a promoter region) of ProDH to identify cis-acting elements involved in L-Pro-induced and hypoosmolarity-induced expression in transgenic tobacco (Nicotiana tabacum) and Arabidopsis plants. We found that a 9-bp sequence, ACTCATCCT, in the ProDH promoter is necessary for the efficient expression of ProDH in response to L-Pro and hypoosmolarity. Moreover, ACTCAT is a core cis-acting element, which we have called Pro- or hypoosmolarity-responsive element (PRE), that is necessary for L-Pro-responsive and hypoosmolarity-responsive expression of ProDH. Microarray and RNA gel-blot analyses showed that 21 L-Pro-inducible genes have the PRE sequences in their promoter regions. These results indicate that the PRE sequence play an important role in the L-Pro-responsive gene expression.
Figures




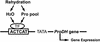
References
-
- Delauney AJ, Hu C-AA, Kishor PBK, Verma DPS. Cloning of ornithine-δ-aminotransferase cDNA from Vigna aconitifolia by trans-complementation in Escherichia coli and regulation of proline biosynthesis. J Biol Chem. 1993;268:18673–18678. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

