Inventory and functional characterization of the HAK potassium transporters of rice
- PMID: 12376644
- PMCID: PMC166606
- DOI: 10.1104/pp.007781
Inventory and functional characterization of the HAK potassium transporters of rice
Abstract
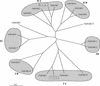
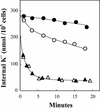
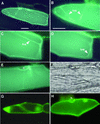
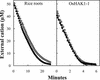
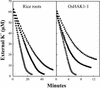
Plants take up large amounts of K(+) from the soil solution and distribute it to the cells of all organs, where it fulfills important physiological functions. Transport of K(+) from the soil solution to its final destination is mediated by channels and transporters. To better understand K(+) movements in plants, we intended to characterize the function of the large KT-HAK-KUP family of transporters in rice (Oryza sativa cv Nipponbare). By searching in databases and cDNA cloning, we have identified 17 genes (OsHAK1-17) encoding transporters of this family and obtained evidence of the existence of other two genes. Phylogenetic analysis of the encoded transporters reveals a great diversity among them, and three distant transporters, OsHAK1, OsHAK7, and OsHAK10, were expressed in yeast (Saccharomyces cerevisiae) and bacterial mutants to determine their functions. The three transporters mediate K(+) influxes or effluxes, depending on the conditions of the experiment. A comparative kinetic analysis of HAK-mediated K(+) influx in yeast and in roots of K(+)-starved rice seedlings demonstrated the involvement of HAK transporters in root K(+) uptake. We discuss that all HAK transporters may mediate K(+) transport, but probably not only in the plasma membrane. Transient expression of the OsHAK10-green fluorescent protein fusion protein in living onion epidermal cells targeted this protein to the tonoplast.
Figures

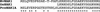
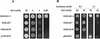
References
-
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. - PubMed
-
- Bakker EP. Low-affinity K+uptake systems. In: Bakker EP, editor. Alkali Cation Transport Systems in Prokaryotes. Boca Raton, FL: CRC Press; 1993. pp. 253–276.
-
- Bañuelos MA, Madrid R, Rodríguez-Navarro A. Individual functions of the HAK and TRK potassium transporters of Schwanniomyces occidentalis. Mol Microbiol. 2000;37:671–679. - PubMed
-
- Barbier-Brygoo H, Vinauger M, Colcombet J, Ephritikhine G, Frachisse J-M, Maurel C. Anion channels in higher plants: functional characterization, molecular structure and physiological role. Biochim Biophys Acta. 2000;1465:199–218. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

