SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis
- PMID: 12376646
- PMCID: PMC166608
- DOI: 10.1104/pp.003491
SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis
Abstract
The importance of maternal cells in controlling early embryogenesis is well understood in animal development, yet in plants the precise role of maternal cells in embryogenesis is unclear. We demonstrated previously that maternal activity of the SIN1 (SHORT INTEGUMENTS1) gene of Arabidopsis is essential for embryo pattern formation and viability, and that its postembryonic activity is required for several processes in reproductive development, including flowering time control and ovule morphogenesis. Here, we report the cloning of SIN1, and demonstrate its identity to the CAF (CARPEL FACTORY) gene important for normal flower morphogenesis and to the SUS1 (SUSPENSOR1) gene essential for embryogenesis. SIN1/SUS1/CAF has sequence similarity to the Drosophila melanogaster gene Dicer, which encodes a multidomain ribonuclease specific for double-stranded RNA, first identified by its role in RNA silencing. The Dicer protein is essential for temporal control of development in animals, through the processing of small RNA hairpins that in turn inhibit the translation of target mRNAs. Structural modeling of the wild-type and sin1 mutant proteins indicates that the RNA helicase domain of SIN1/SUS1/CAF is important for function. The mRNA was detected in floral meristems, ovules, and early embryos, consistent with the mutant phenotypes. A 3.3-kb region 5' of the SIN1/SUS1/CAF gene shows asymmetric parent-of-origin activity in the embryo: It confers transcriptional activation of a reporter gene in early embryos only when transmitted through the maternal gamete. These results suggest that maternal SIN1/SUS1/CAF functions early in Arabidopsis development, presumably through posttranscriptional regulation of specific mRNA molecules.
Figures

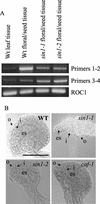



References
-
- Baroux C, Spillane C, Grossniklaus U. Genomic imprinting during seed development. In: Dunlap JC, Wu C-t, editors. Advances in Genetics: Homology Effects. San Diego: Academic Press; 2002. pp. 165–214. - PubMed
-
- Bernstein E, Caudy A, Hammond S, Hannon G. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

