Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato
- PMID: 12376655
- PMCID: PMC166617
- DOI: 10.1104/pp.007427
Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato
Abstract
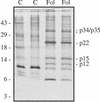
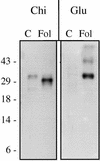
The protein content of tomato (Lycopersicon esculentum) xylem sap was found to change dramatically upon infection with the vascular wilt fungus Fusarium oxysporum. Peptide mass fingerprinting and mass spectrometric sequencing were used to identify the most abundant proteins appearing during compatible or incompatible interactions. A new member of the PR-5 family was identified that accumulated early in both types of interaction. Other pathogenesis-related proteins appeared in compatible interactions only, concomitantly with disease development. This study demonstrates the feasibility of using proteomics for the identification of known and novel proteins in xylem sap, and provides insights into plant-pathogen interactions in vascular wilt diseases.
Figures





References
-
- Abad L, D'Urzo MP, Liu D, Narasimhan ML, Reuveni M, Zhu JK, Niu X, Singh NK, Hasegawa PM, Bressan RA. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996;118:11–23.
-
- Beckman CH, Roberts EM. On the nature and genetic basis for resistance and tolerance to fungal wilt diseases of plants. Adv Bot Res. 1995;21:35–77.
-
- Benhamou N, Grenier J, Asselin A. Immunogold localization of pathogenesis-related protein P14 in tomato root cells infected with Fusarium oxysporum f. sp. radicis-lycopersici. Physiol Mol Plant Pathol. 1991;38:237–253.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

