Genetic architecture of NaCl tolerance in Arabidopsis
- PMID: 12376659
- PMCID: PMC166621
- DOI: 10.1104/pp.006536
Genetic architecture of NaCl tolerance in Arabidopsis
Abstract
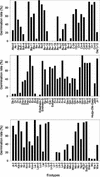
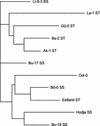
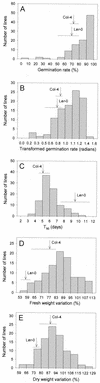
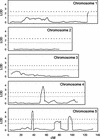
The little success of breeding approaches toward the improvement of salt tolerance in crop species is thought to be attributable to the quantitative nature of most, if not all the processes implicated. Hence, the identification of some of the quantitative trait loci (QTL) that contribute to natural variation in salt tolerance should be instrumental in eventually manipulating the perception of salinity and the corresponding responses. A good choice to reach this goal is the plant model system Arabidopsis, whose complete genome sequence is now available. Aiming to analyze natural variability in salt tolerance, we have compared the ability of 102 wild-type races (named ecotypes or accessions) of Arabidopsis to germinate on 250 mM NaCl, finding a wide range of variation among them. Accessions displaying extremely different responses to NaCl were intercrossed, and the phenotypes found in their F(2) progenies suggested that natural variation in NaCl tolerance during germination was under polygenic controls. Genetic distances calculated on the basis of variations in repeat number at 22 microsatellites, were analyzed in a group of either extremely salt-tolerant or extremely salt-sensitive accessions. We found that most but not all accessions with similar responses to NaCl are phylogenetically related. NaCl tolerance was also studied in 100 recombinant inbred lines derived from a cross between the Columbia-4 and Landsberg erecta accessions. We detected 11 QTL harboring naturally occurring alleles that contribute to natural variation in NaCl tolerance in Arabidopsis, six at the germination and five at the vegetative growth stages, respectively. At least five of these QTL are likely to represent loci not yet described by their relationship with salt stress.
Figures

References
-
- Abel GH, Mackenzie AJ. Salt tolerance of soybean varieties (Glycine max L. Merrill) during germination and later growth. Crop Sci. 1963;3:159–161.
-
- Alonso-Blanco C, Koornneef M. Naturally occurring variation in Arabidopsis: an underexploited resource for plants genetics. Trends Plant Sci. 2000;5:22–29. - PubMed
-
- Barkla BJ, Vera-Estrella R, Pantoja O. Towards the production of salt-tolerant crops. In: Shadidi F, Kolodziejczyk P, Whitaker JR, Munguia AL, Fuller G, editors. Chemicals via Higher Plant Bioengineering. 464 of Advances in Experimental Medicine and Biology. New York: Kluwer Academic/Plenum Publishers; 1999. pp. 77–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

