Contributions of the N- and C-terminal domains of surfactant protein d to the binding, aggregation, and phagocytic uptake of bacteria
- PMID: 12379690
- PMCID: PMC130308
- DOI: 10.1128/IAI.70.11.6129-6139.2002
Contributions of the N- and C-terminal domains of surfactant protein d to the binding, aggregation, and phagocytic uptake of bacteria
Abstract
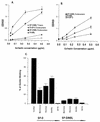
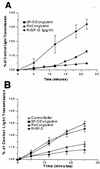
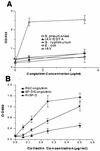
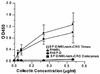
Collectins play important roles in host defense against infectious microorganisms. We now demonstrate that the serum collectins mannose-binding lectin (MBL) and conglutinin have less ability to bind to, aggregate, and enhance neutrophil uptake of several strains of gram-negative and gram-positive bacteria than pulmonary surfactant protein D (SP-D). Collectins are composed of four major structural domains (i.e., N-terminal, collagen, and neck and carbohydrate recognition domains). To determine which domains of SP-D are responsible for its greater bacterial binding or aggregating activity, activities of chimeric collectins containing the N-terminal and collagen domains of SP-D coupled to the neck recognition domains and carbohydrate recognition domains (CRD) of MBL or conglutinin (SP-D/Cong(neck+CRD) and SP-D/MBL(neck+CRD)) were tested. The SP-D/Cong(neck+CRD) and SP-D/MBL(neck+CRD) chimeras bound to and aggregated the bacteria more strongly than did wild-type MBL or conglutinin. SP-D/MBL(neck+CRD) also enhanced neutrophil uptake of bacteria more so than MBL. Hence, the SP-D N-terminal and/or collagen domains contribute to the enhanced bacterial binding and aggregating activities of SP-D. In prior studies, SP-D/Cong(neck+CRD) and SP-D/MBL(neck+CRD) had increased ability to bind to influenza virus compared not only with that of conglutinin or MBL but with that of wild-type SP-D as well. In contrast, the chimeras had either reduced or unchanged ability to bind to or aggregate bacteria compared to that of wild-type SP-D. Hence, although replacement of the neck recognition domains and CRDs of SP-D with those of MBL and conglutinin conferred increased viral binding activity, it did not favorably affect bacterial binding activity, suggesting that requirements for optimal collectin binding to influenza virus and bacteria differ.
Figures




References
-
- Allen, M., A. Laederach, P. Reilly, and R. Mason. 2001. Polysaccharide recognition by surfactant protein D: novel interactions of a C-type lectin with nonterminal glucosyl residues. Biochemistry 40:7789-7798. - PubMed
-
- Brown-Augsburger, P., D. Chang, K. Rust, and E. Crouch. 1996. Biosynthesis of surfactant protein D. J. Biol. Chem. 271:18912-18919. - PubMed
-
- Crouch, E., D. Chang, K. Rust, A. Persson, and J. Heuser. 1994. Recombinant pulmonary surfactant protein D. J. Biol. Chem. 269:15808-15813. - PubMed
-
- Crouch, E., K. Hartshorn, and I. Ofek. 2000. Collectins and pulmonary innate immunity. Immunol. Rev. 173:52-65. - PubMed
-
- Devyatyarova-Johnson, M., I. Rees, B. Robertson, M. Turner, N. Klein, and D. Jack. 2000. The lipopolysaccharide structures of Salmonella enterica serovar Typhimurium and Neisseria gonorrhoeae determine the attachment of mannose binding lectin to intact organisms. Infect. Immun. 68:3894-3899. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

