Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536
- PMID: 12379716
- PMCID: PMC130402
- DOI: 10.1128/IAI.70.11.6365-6372.2002
Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536
Abstract
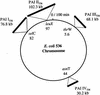
For the uropathogenic Escherichia coli strain 536 (O6:K15:H31), the DNA sequences of three pathogenicity islands (PAIs) (PAI I(536) to PAI III(536)) and their flanking regions (about 270 kb) were determined to further characterize the virulence potential of this strain. PAI I(536) to PAI III(536) exhibit features typical of PAIs, such as (i) association with tRNA-encoding genes; (ii) G+C content differing from that of the host genome; (iii) flanking repeat structures; (iv) a mosaic-like structure comprising a multitude of functional, truncated, and nonfunctional putative open reading frames (ORFs) with known or unknown functions; and (v) the presence of many fragments of mobile genetic elements. PAI I(536) to PAI III(536) range between 68 and 102 kb in size. Although these islands contain several ORFs and known virulence determinants described for PAIs of other extraintestinal pathogenic E. coli (ExPEC) isolates, they also consist of as-yet-unidentified ORFs encoding putative virulence factors. The genetic structure of PAI IV(536), which represents the core element of the so-called high-pathogenicity island encoding a siderophore system initially identified in pathogenic yersiniae, was further characterized by sample sequencing. For the first time, multiple PAI sequences (PAI I(536) to PAI IV(536)) in uropathogenic E. coli were studied and their presence in several wild-type E. coli isolates was extensively investigated. The results obtained suggest that these PAIs or at least large fragments thereof are detectable in other pathogenic E. coli isolates. These results support our view that the acquisition of large DNA regions, such as PAIs, by horizontal gene transfer is an important factor for the evolution of bacterial pathogens.
Figures

References
-
- Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67:4851-4861. - PMC - PubMed
-
- Cieslewicz, M., and E. Vimr. 1997. Reduced polysialic acid capsule expression in Escherichia coli K1 mutants with chromosomal defects in kpsF. Mol. Microbiol. 26:237-249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

