Structural and functional variation within the alanine-rich repetitive domain of streptococcal antigen I/II
- PMID: 12379719
- PMCID: PMC130335
- DOI: 10.1128/IAI.70.11.6389-6398.2002
Structural and functional variation within the alanine-rich repetitive domain of streptococcal antigen I/II
Abstract
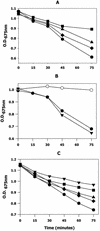
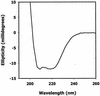
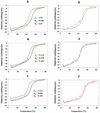
Members of the antigen I/II family of cell surface proteins are highly conserved, multifunctional adhesins that mediate interactions of oral streptococci with other oral bacteria, with cell matrix proteins (e.g., type I collagen), and with salivary glycoproteins, e.g., gp340. The interaction of gp340 (formerly designated salivary agglutinin) with Streptococcus mutans requires an alanine-rich repetitive domain (A region) of antigen I/II that is highly conserved in all members of this family of proteins. In this report, we show that the A regions from the two Streptococcus gordonii M5 antigen I/II proteins (SspA and SspB) interact differently with the salivary gp340 glycoprotein and appear to be structurally distinct. Recombinant polypeptides encompassing the A region of SspA or from a highly related S. mutans antigen I/II protein (SpaP) competitively inhibited the interaction of gp340 with intact S. gordonii and S. mutans cells, respectively. In contrast, an A region polypeptide from SspB was inactive, and furthermore, it did not bind to purified gp340 in vitro. Circular dichroism spectra suggested that all three polypeptides were highly alpha-helical and may form coiled-coil structures. However, the A region of SspB underwent a conformational change and exhibited reduced alpha-helical structure at pH 8.5, whereas the A region polypeptides from SspA and SpaP were relatively stable under these conditions. Melt curves also indicated that at physiological pH, the A region of SspB lost alpha-helical structure more rapidly than that of SspA or SpaP when the temperature was increased from 10 to 40 degrees C. Furthermore, the SspB A region polypeptide denatured completely at a temperature that was 7 to 9 degrees C lower than that required for the A region polypeptide of SspA or SpaP. The full-length SspB protein and the three A region peptides migrated in native gel electrophoresis and column chromatography with apparent molecular masses that were approximately 2- to 2.5-fold greater than their predicted molecular masses. However, sedimentation equilibrium ultracentrifugation data showed that the A region peptides sedimented as monomers, suggesting that the peptides may form nonglobular intramolecular coiled-coil structures under the experimental conditions used. Taken together, our results suggest that the A region of SspB is less stable than the corresponding A regions of SspA and SpaP and that this structural difference may explain, at least in part, the functional variation observed in their interactions with salivary gp340.
Figures

Similar articles
-
Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands.Mol Microbiol. 2005 Mar;55(5):1591-605. doi: 10.1111/j.1365-2958.2005.04495.x. Mol Microbiol. 2005. PMID: 15720563
-
Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363.Infect Immun. 1998 Oct;66(10):4633-9. doi: 10.1128/IAI.66.10.4633-4639.1998. Infect Immun. 1998. PMID: 9746559 Free PMC article.
-
Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors.Infect Immun. 2005 Oct;73(10):6629-38. doi: 10.1128/IAI.73.10.6629-6638.2005. Infect Immun. 2005. PMID: 16177339 Free PMC article.
-
The changing faces of Streptococcus antigen I/II polypeptide family adhesins.Mol Microbiol. 2010 Jul;77(2):276-86. doi: 10.1111/j.1365-2958.2010.07212.x. Epub 2010 May 24. Mol Microbiol. 2010. PMID: 20497507 Free PMC article. Review.
-
Structure, function and immunogenicity of streptococcal antigen I/II polypeptides.Mol Microbiol. 1997 Jan;23(2):183-90. doi: 10.1046/j.1365-2958.1997.2021577.x. Mol Microbiol. 1997. PMID: 9044252 Review.
Cited by
-
Identification and genetic characterization of a novel proteinase, PrtR, from the human isolate Lactobacillus rhamnosus BGT10.Appl Environ Microbiol. 2003 Oct;69(10):5802-11. doi: 10.1128/AEM.69.10.5802-5811.2003. Appl Environ Microbiol. 2003. PMID: 14532028 Free PMC article.
-
Redirecting the humoral immune response against Streptococcus mutans antigen P1 with monoclonal antibodies.Infect Immun. 2004 Dec;72(12):6951-60. doi: 10.1128/IAI.72.12.6951-6960.2004. Infect Immun. 2004. PMID: 15557617 Free PMC article.
-
Molecular interactions of surface protein peptides of Streptococcus gordonii with human salivary components.Infect Immun. 2004 Aug;72(8):4819-26. doi: 10.1128/IAI.72.8.4819-4826.2004. Infect Immun. 2004. PMID: 15271944 Free PMC article.
-
Identification of glucosyl transferase inhibitors from Psidium guajava against Streptococcus mutans in dental caries.J Tradit Complement Med. 2018 May 1;9(2):124-137. doi: 10.1016/j.jtcme.2017.09.003. eCollection 2019 Apr. J Tradit Complement Med. 2018. PMID: 30963047 Free PMC article.
References
-
- Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism with an unsupervised neural network. Protein Eng. 6:383-390. - PubMed
-
- Bikker, F. J., A. J. Ligtenberg, J. E. van der Wal, P. A. van den Keijbus, U. Holmskov, E. C. Veerman, and A. V. Nieuw Amerongen. 2002. Immunohistochemical detection of salivary agglutinin/gp340 in human parotid, submandibular, and labial salivary glands. J. Dent. Res. 81:134-139. - PubMed
-
- Brady, L. J., D. G. Cvitkovitch, C. M. Geric, M. N. Addison, J. C. Joyce, P. J. Crowley, and A. S. Bleiweis. 1998. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect. Immun.. 66:4274-4282. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

