MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison
- PMID: 12381844
- PMCID: PMC2373724
- DOI: 10.1110/ps.0215902
MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison
Abstract
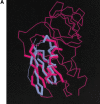
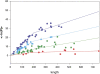
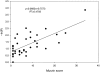
Advances in structural genomics and protein structure prediction require the design of automatic, fast, objective, and well benchmarked methods capable of comparing and assessing the similarity of low-resolution three-dimensional structures, via experimental or theoretical approaches. Here, a new method for sequence-independent structural alignment is presented that allows comparison of an experimental protein structure with an arbitrary low-resolution protein tertiary model. The heuristic algorithm is given and then used to show that it can describe random structural alignments of proteins with different folds with good accuracy by an extreme value distribution. From this observation, a structural similarity score between two proteins or two different conformations of the same protein is derived from the likelihood of obtaining a given structural alignment by chance. The performance of the derived score is then compared with well established, consensus manual-based scores and data sets. We found that the new approach correlates better than other tools with the gold standard provided by a human evaluator. Timings indicate that the algorithm is fast enough for routine use with large databases of protein models. Overall, our results indicate that the new program (MAMMOTH) will be a good tool for protein structure comparisons in structural genomics applications. MAMMOTH is available from our web site at http://physbio.mssm.edu/~ortizg/.
Figures









References
-
- Abagyan, R. and Batalov, S. 1997. Do aligned sequences share the same fold? J. Mol. Biol. 273 355–368. - PubMed
-
- Adams, P.D. and Grosse-Kunstleve, R.W. 2000. Recent developments in software for the automation of crystallographic macromolecular structure determination. Curr. Opin. Struct. Biol. 10 564–568. - PubMed
-
- Al-Hashimi, H.M. and Patel, D.J. 2002. Residual dipolar couplings: Synergy between NMR and structural genomics. J. Biomol. NMR 22 1–8. - PubMed
-
- Baker, D. and Sali, A. 2001. Protein structure prediction and structural genomics. Science 294 93–96. - PubMed
-
- Bonneau, B., Strauss, C., Rohl, C., Chivian, D., Bradley, P., Malmstrom, L., Robertson, T., Baker, D. 2002. De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 322 65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

