A statistically derived parameterization for the collagen triple-helix
- PMID: 12381857
- PMCID: PMC2373720
- DOI: 10.1110/ps.0218502
A statistically derived parameterization for the collagen triple-helix
Erratum in
- Protein Sci. 2004 Aug;13(8):2276
Abstract
The triple-helix is a unique secondary structural motif found primarily within the collagens. In collagen, it is a homo- or hetero-tripeptide with a repeating primary sequence of (Gly-X-Y)(n), displaying characteristic peptide backbone dihedral angles. Studies of bulk collagen fibrils indicate that the triple-helix must be a highly repetitive secondary structure, with very specific constraints. Primary sequence analysis shows that most collagen molecules are primarily triple-helical; however, no high-resolution structure of any entire protein is yet available. Given the drastic morphological differences in self-assembled collagen structures with subtle changes in assembly conditions, a detailed knowledge of the relative locations of charged and sterically bulky residues in collagen is desirable. Its repetitive primary sequence and highly conserved secondary structure make collagen, and the triple-helix in general, an ideal candidate for a general parameterization for prediction of residue locations and for the use of a helical wheel in the prediction of residue orientation. Herein, a statistical analysis of the currently available high-resolution X-ray crystal structures of model triple-helical peptides is performed to produce an experimentally based parameter set for predicting peptide backbone and C(beta) atom locations for the triple-helix. Unlike existing homology models, this allows easy prediction of an entire triple-helix structure based on all existing high-resolution triple-helix structures, rather than only on a single structure or on idealized parameters. Furthermore, regional differences based on the helical propensity of residues may be readily incorporated. The parameter set is validated in terms of the predicted bond lengths, backbone dihedral angles, and interchain hydrogen bonding.
Figures
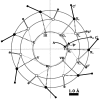
References
-
- Barnett, V. and Lewis, T. 1994. Outliers in statistical data, 3rd ed. John Wiley & Sons, Chichester, United Kingdom.
-
- Beck, K. and Brodsky, B. 1998. Supercoiled protein motifs: The collagen triple-helix and the α-helical coiled coil. J. Struct. Biol. 122 17–29. - PubMed
-
- Bella, J., Eaton, M., Brodsky, B., and Berman, H.M. 1994. Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science 266 75–81. - PubMed
-
- Bella, J., Brodsky, B., and Berman, H.M. 1995. Hydration structure of a collagen peptide. Structure 3 893–906. - PubMed
Web references
-
- http://www.bioinf.org.uk/software/profit; ProFit 2.2.
-
- http://www.rcsb.org/pdb; Protein Data Bank.
-
- http://www.fccc.edu/research/labs/dunbrack/scwrl; SCWRL 2.95.
-
- http://www.chem.utoronto.ca/staff/MCG/; Web-based interface for the generation of triple-helical structures of any primary sequence and length.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

