Does RNA polymerase help drive chromosome segregation in bacteria?
- PMID: 12384568
- PMCID: PMC137841
- DOI: 10.1073/pnas.182539899
Does RNA polymerase help drive chromosome segregation in bacteria?
Abstract
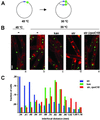
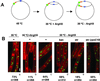
In contrast to eukaryotic cells, bacteria segregate their chromosomes without a conspicuous mitotic apparatus. Replication of bacterial chromosomes initiates bidirectionally from a single site (oriC), and visualization of the region of the chromosome containing oriC in living cells reveals that origins rapidly move apart toward opposite poles of the cell during the cell cycle. The motor that drives this poleward movement is unknown. An attractive candidate is RNA polymerase, which is a powerful and abundant molecular motor. If, as has been suggested for other macromolecular complexes, the movement of RNA polymerase is restricted in the cell, then transcription would translocate the DNA template, thereby providing the motive force to separate replicating chromosomes. A coordinated effect of many transcribing RNA polymerases could result from the widely conserved global bias of gene orientation away from oriC. By using fluorescence microscopy of living Bacillus subtilis cells, we demonstrate that an inhibitor of RNA polymerase acts to inhibit separation of newly duplicated DNAs near the origin of replication. We propose that the force exerted by RNA polymerase contributes to chromosome movement in bacteria, and that this force, coupled with the biased orientation of transcription units, helps to drive chromosome segregation.
Figures

References
-
- Jacob F., Brenner, S. & Cuzin, F. (1963) Cold Spring Harbor Symp. Quant. Biol. 23, 329-348.
-
- Webb C. D., Graumann, P. L., Kahana, J. A., Teleman, A. A., Silver, P. A. & Losick, R. (1998) Mol. Microbiol. 28, 883-892. - PubMed
-
- Webb C. D., Teleman, A., Gordon, S., Straight, A., Belmont, A., Lin, D. C., Grossman, A. D., Wright, A. & Losick, R. (1997) Cell 88, 667-674. - PubMed
-
- Glaser P., Sharpe, M. E., Raether, B., Perego, M., Ohlsen, K. & Errington, J. (1997) Genes Dev. 11, 1160-1168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

