Biochemical characterisation of the clamp/clamp loader proteins from the euryarchaeon Archaeoglobus fulgidus
- PMID: 12384579
- PMCID: PMC137147
- DOI: 10.1093/nar/gkf584
Biochemical characterisation of the clamp/clamp loader proteins from the euryarchaeon Archaeoglobus fulgidus
Abstract
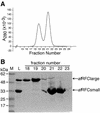
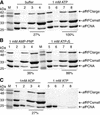
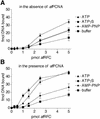
Replicative polymerases of eukaryotes, prokaryotes and archaea obtain processivity using ring-shaped DNA sliding clamps that are loaded onto DNA by clamp loaders [replication factor C (RFC) in eukaryotes]. In this study, we cloned the two genes for the subunits of the RFC homologue of the euryarchaeon Archaeoglobus fulgidus. The proteins were expressed and purified from Escherichia coli both individually and as a complex. The afRFC subunits form a heteropentameric complex consisting of one copy of the large subunit and four copies of the small subunits. To analyse the functionality of afRFC, we also expressed the A.fulgidus PCNA homologue and a type B polymerase (PolB1) in E.coli. In primer extension assays, afRFC stimulated the processivity of afPolB1 in afPCNA-dependent reactions. Although the afRFC complex showed significant DNA-dependent ATPase activity, which could be further stimulated by afPCNA, neither of the isolated afRFC subunits showed this activity. However, both the large and small afRFC subunits showed interaction with afPCNA. Furthermore, we demonstrate that ATP binding, but not hydrolysis, is needed to stimulate interactions of the afRFC complex with afPCNA and DNA.
Figures




References
-
- Jeruzalmi D., O’Donnell,M. and Kuriyan,J. (2002) Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol., 12, 217–224. - PubMed
-
- Onrust R. and O’Donnell,M. (1993) DNA polymerase III accessory proteins. II. Characterization of δ and δ′. J. Biol. Chem., 268, 11766–11772. - PubMed
-
- Jeruzalmi D., O’Donnell,M. and Kuriyan,J. (2001) Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell, 106, 429–441. - PubMed
-
- Jeruzalmi D., Yurieva,O., Zhao,Y., Young,M., Stewart,J., Hingorani,M., O’Donnell,M. and Kuriyan,J. (2001) Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell, 106, 417–428. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

