Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots
- PMID: 12388687
- PMCID: PMC136766
- DOI: 10.1128/jvi.76.22.11273-11282.2002
Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots
Abstract
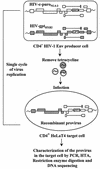
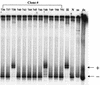
Previously, we reported that human immunodeficiency virus type 1 (HIV-1) recombines approximately two to three times per genome per replication cycle, an extremely high rate of recombination given the relatively small genome size of HIV-1. However, a recombination hot spot involving sequence of nonretroviral origin was identified in the vector system utilized, raising the possibility that this hot spot skewed the rate of recombination, and the rate of recombination observed was an overestimation. To address this issue, an HIV-1-derived vector system was used to examine the rate of recombination between autologous HIV-1 sequences after restricting replication to a single cycle in the absence of this hot spot. Viral DNA and RNA were analyzed by a combination of the heteroduplex tracking assay, restriction enzyme analysis, DNA sequencing, and reverse transcription-PCR. The results indicate that HIV-1 undergoes recombination at a minimum rate of 2.8 crossovers per genome per cycle. Again, this is a very high rate given the small size of the HIV-1 genome. The results also suggested that there might be local hot spots of recombination at different locations throughout the genome since 13 of the 33 strand transfers identified by DNA sequencing shared the same site of recombination with one or two other clones. Furthermore, identification of crossover segments also allowed examination of mutations at the point of recombination, since it has been predicted from some studies of cell-free systems that mutations may occur with a frequency of 30 to 50% at crossover junctions. However, DNA sequence analysis of crossover junctions indicated that homologous recombination during viral replication was not particularly mutagenic, indicating that there are other factors or conditions not yet reproduced in cell-free systems which contribute to fidelity during retroviral recombination.
Figures




References
-
- Balakrishnan, M., P. J. Fay, and R. A. Bambara. 2001. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 276:36482-36492. - PubMed
-
- Battula, N., and L. A. Loeb. 1976. On the fidelity of DNA replication. Lack of exodeoxyribonuclease activity and error-correcting function in avian myeloblastosis virus DNA polymerase. J. Biol. Chem. 251:982-986. - PubMed
-
- Bebenek, K., J. Abbotts, J. D. Roberts, S. H. Wilson, and T. A. Kunkel. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 264:16948-16956. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

