Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus
- PMID: 12388706
- PMCID: PMC136748
- DOI: 10.1128/jvi.76.22.11447-11459.2002
Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus
Erratum in
- J Virol. 2003 Mar;77(5):3351.
Abstract
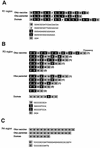
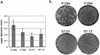
The DNA sequences of the Oka varicella vaccine virus (V-Oka) and its parental virus (P-Oka) were completed. Comparison of the sequences revealed 42 base substitutions, which led to 20 amino acid conversions and length differences in tandem repeat regions (R1, R3, and R4) and in an origin of DNA replication. Amino acid substitutions existed in open reading frames (ORFs) 6, 9A, 10, 21, 31, 39, 50, 52, 55, 59, 62, and 64. Of these, 15 base substitutions, leading to eight amino acid substitutions, were in the gene 62 region alone. Further DNA sequence analysis showed that these substitutions were specific for V-Oka and were not present in nine clinical isolates. The immediate-early gene 62 product (IE62) of P-Oka had stronger transactivational activity than the mutant IE62 contained in V-Oka in 293 and CV-1 cells. An infectious center assay of a plaque-purified clone (S7-01) from the V-Oka with 8 amino acid substitutions in ORF 62 showed smaller plaque formation and less-efficient virus-spreading activity than did P-Oka in human embryonic lung cells. Another clone (S-13) with only five substitutions in ORF 62 spread slightly faster than S7-01 but not as effectively as P-Oka. Moreover, transient luciferase assay in 293 cells showed that transactivational activities of IE62s of S7-01 and S7-13 were lower than that of P-Oka. Based on these results, it appears that amino acid substitutions in ORF 62 are responsible for virus growth and spreading from infected to uninfected cells. Furthermore, the Oka vaccine virus was completely distinguishable from P-Oka and 54 clinical isolates by seven restriction-enzyme fragment length polymorphisms that detected differences in the DNA sequence.
Figures



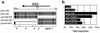
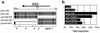


References
-
- Argaw, T., J. I. Cohen, M. Klutch, K. Lekstrom, T. Yoshikawa, Y. Asano, and P. R. Krause. 2000. Nucleotide sequences that distinguish Oka vaccine from parental Oka and other varicella-zoster virus isolates. J. Infect. Dis. 181:1153-1157. - PubMed
-
- Asano, Y., N. Itakura, Y. Hiroishi, S. Hirose, T. Ozaki, K. Kuno, T. Nagai, T. Yazaki, K. Yamanishi, and M. Takahashi. 1985. Viral replication and immunologic responses in children naturally infected with varicella-zoster virus and in varicella vaccine recipients. J. Infect. Dis. 152:863-868. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

