The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells
- PMID: 12388727
- PMCID: PMC136756
- DOI: 10.1128/jvi.76.22.11677-11687.2002
The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells
Abstract
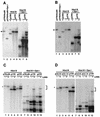
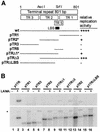
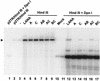
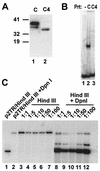
Kaposi's sarcoma-associated herpesvirus (KSHV) is associated with Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. The latency-associated nuclear antigen (LANA) is a multifunctional protein that is consistently expressed in all KSHV-associated malignancies. LANA interacts with a variety of cellular proteins, including the transcriptional cosuppressor complex mSin3 and the tumor suppressors p53 and Rb, thereby regulating viral and cellular gene expression. In addition, LANA is required for maintenance of the episomal viral DNA during latency in dividing cells. Colocalization studies suggest that LANA tethers the viral genome to chromosomes during mitosis. In support of this model, a specific LANA- binding site has recently been identified within the terminal repeat unit, and a chromatin interaction domain was mapped to a short amino acid stretch within the N-terminal domain of LANA. Epstein-Barr virus nuclear antigen 1 (EBNA-1), a functional homologue of LANA, is also required for genome segregation; in addition, EBNA-1 also supports efficient DNA replication of oriP-containing plasmids. By performing short-term replication assays, we demonstrate here for the first time that de novo synthesis of terminal-repeat (TR)-containing plasmids is highly dependent on the presence of LANA. We map the required cis-acting sequences within the TR to a 79-bp region and demonstrate that the DNA-binding domain of LANA is required for this DNA replication activity. Surprisingly, the 233-amino-acid C domain of LANA by itself partially supports replication. Our data show that LANA is a sequence-specific DNA-binding protein that, like EBNA-1, plays an important role in DNA replication and genome segregation. In addition, we show that all necessary cis elements for the origin of replication (ori) function are located within a single TR, suggesting that the putative ori of KSHV is different from those of other gammaherpesviruses, which all contain ori sequences within the unique long sequence outside of their TR. This notion is further strengthened by the unique modular structure of the KSHV TR element.
Figures

Similar articles
-
DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus.J Virol. 2001 Sep;75(17):7882-92. doi: 10.1128/jvi.75.17.7882-7892.2001. J Virol. 2001. PMID: 11483733 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus LANA-Adjacent Regions with Distinct Functions in Episome Segregation or Maintenance.J Virol. 2019 Mar 5;93(6):e02158-18. doi: 10.1128/JVI.02158-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626680 Free PMC article.
-
Minichromosome Maintenance Proteins Cooperate with LANA during the G1/S Phase of the Cell Cycle To Support Viral DNA Replication.J Virol. 2019 Mar 21;93(7):e02256-18. doi: 10.1128/JVI.02256-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651368 Free PMC article.
-
KSHV Genome Replication and Maintenance in Latency.Adv Exp Med Biol. 2018;1045:299-320. doi: 10.1007/978-981-10-7230-7_14. Adv Exp Med Biol. 2018. PMID: 29896673 Review.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
Cited by
-
The RBP-Jκ binding sites within the RTA promoter regulate KSHV latent infection and cell proliferation.PLoS Pathog. 2012 Jan;8(1):e1002479. doi: 10.1371/journal.ppat.1002479. Epub 2012 Jan 12. PLoS Pathog. 2012. PMID: 22253595 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus-encoded LANA recruits topoisomerase IIβ for latent DNA replication of the terminal repeats.J Virol. 2012 Sep;86(18):9983-94. doi: 10.1128/JVI.00839-12. Epub 2012 Jul 3. J Virol. 2012. PMID: 22761383 Free PMC article.
-
Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1.J Virol. 2005 Nov;79(21):13618-29. doi: 10.1128/JVI.79.21.13618-13629.2005. J Virol. 2005. PMID: 16227282 Free PMC article.
-
Crystal structure of the gamma-2 herpesvirus LANA DNA binding domain identifies charged surface residues which impact viral latency.PLoS Pathog. 2013;9(10):e1003673. doi: 10.1371/journal.ppat.1003673. Epub 2013 Oct 17. PLoS Pathog. 2013. PMID: 24146618 Free PMC article.
-
Plasmid Partitioning by Human Tumor Viruses.J Virol. 2018 Apr 13;92(9):e02170-17. doi: 10.1128/JVI.02170-17. Print 2018 May 1. J Virol. 2018. PMID: 29467315 Free PMC article. Review.
References
-
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

