Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag
- PMID: 12388733
- PMCID: PMC136778
- DOI: 10.1128/jvi.76.22.11729-11737.2002
Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag
Abstract
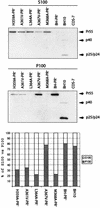
A 14-amino-acid spacer peptide termed SP1 that separates the capsid (CA) and nucleocapsid (NC) sequences plays an active role in the assembly of human immunodeficiency virus type 1. This activity of SP1 involves its amino-terminal residues that, together with adjacent CA residues, constitute a putative alpha-helical structure spanning Gag residues from positions 359 to 371. In this study, we have determined that the virus assembly determinants within this putative alpha-helix were residues H359, K360, A361, L364, A367, and M368, of which K360 and A367 contribute to virus production to lesser extents. Notably, changes of the two basic amino acids H359 and K360 to arginine (R) impaired virus production, whereas mutations L364I and M368I, in contrast to L364A and M368A, generated near-wild-type levels of virus particles. This suggests that within Gag complexes, amino acids H359 and K360 are involved in stricter steric interactions than L364 and M368. Since L364 and M368 are separated by four residues and thus presumably located on the same side of the helical surface, they may initiate synergistic hydrophobic interactions to stabilize Gag association. Further analysis in the context of the protease-negative mutation D185H confirmed the key roles of amino acids H359, A361, L364, and M368 in virus assembly. Importantly, when transfected cells were subjected to Dounce homogenization and the cell lysates were treated by ultracentrifugation at 100,000 x g, Gag molecules containing each of the H359A, A361V, L364A, and M368A mutations were found mainly in the supernatant fraction (S100), whereas approximately 80% of wild-type Gag proteins were found in the pellet. Therefore, these four mutations must have prevented Gag from generating large complexes.
Figures




Similar articles
-
Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag.J Virol. 2005 Feb;79(3):1803-12. doi: 10.1128/JVI.79.3.1803-1812.2005. J Virol. 2005. PMID: 15650204 Free PMC article.
-
Opposing effects of the M368A point mutation and deletion of the SP1 region on membrane binding of human immunodeficiency virus type 1 Gag.Virology. 2005 May 10;335(2):232-41. doi: 10.1016/j.virol.2005.03.004. Virology. 2005. PMID: 15840522
-
Important role for the CA-NC spacer region in the assembly of bovine immunodeficiency virus Gag protein.J Virol. 2004 Jan;78(2):551-60. doi: 10.1128/jvi.78.2.551-560.2004. J Virol. 2004. PMID: 14694086 Free PMC article.
-
The HIV-1 assembly machine.AIDS. 2001;15 Suppl 5:S13-20. doi: 10.1097/00002030-200100005-00003. AIDS. 2001. PMID: 11816161 Review. No abstract available.
-
Stephan Oroszlan and the Proteolytic Processing of Retroviral Proteins: Following A Pro.Viruses. 2021 Nov 4;13(11):2218. doi: 10.3390/v13112218. Viruses. 2021. PMID: 34835024 Free PMC article. Review.
Cited by
-
CryoET structures of immature HIV Gag reveal six-helix bundle.Commun Biol. 2021 Apr 16;4(1):481. doi: 10.1038/s42003-021-01999-1. Commun Biol. 2021. PMID: 33863979 Free PMC article.
-
Structural and functional insights into the HIV-1 maturation inhibitor binding pocket.PLoS Pathog. 2012;8(11):e1002997. doi: 10.1371/journal.ppat.1002997. Epub 2012 Nov 8. PLoS Pathog. 2012. PMID: 23144615 Free PMC article.
-
Incorporation of human immunodeficiency virus type 1 reverse transcriptase into virus-like particles.J Virol. 2007 May;81(10):5155-65. doi: 10.1128/JVI.01796-06. Epub 2007 Mar 7. J Virol. 2007. PMID: 17344306 Free PMC article.
-
Relationships between infectious titer, capsid protein levels, and reverse transcriptase activities of diverse human immunodeficiency virus type 1 isolates.J Virol. 2004 Oct;78(20):11130-41. doi: 10.1128/JVI.78.20.11130-11141.2004. J Virol. 2004. PMID: 15452233 Free PMC article.
-
The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization.J Virol. 2007 Jun;81(12):6216-30. doi: 10.1128/JVI.00284-07. Epub 2007 Apr 11. J Virol. 2007. PMID: 17428849 Free PMC article.
References
-
- Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

