Mutation of capsid protein phosphorylation sites abolishes cauliflower mosaic virus infectivity
- PMID: 12388736
- PMCID: PMC136793
- DOI: 10.1128/jvi.76.22.11748-11752.2002
Mutation of capsid protein phosphorylation sites abolishes cauliflower mosaic virus infectivity
Abstract
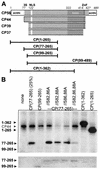
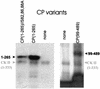
The cauliflower mosaic virus (CaMV) capsid protein is derived by bidirectional processing of the precapsid protein (CP56). We expressed several derivatives of CP56 in Escherichia coli and used them as substrates for virus-associated kinase and casein kinase II purified from plant cells. Three serine residues located at the N terminus of the mature viral protein CP44 were identified as phosphorylation targets. A mutation of one of them in the viral context had little or no effect on viral infectivity, but a mutation of all three serines abolished infectivity. The mapping of phosphorylation sites in CP44, but not CP39 or CP37, and immunodetection of the Zn finger motif in CP44 and CP39, but not CP37, support the model that CP39 is produced from CP44 by N-terminal processing and CP37 is produced from CP39 by C-terminal processing. We discuss the possible role of phosphorylation in the processing and assembly of CaMV capsid protein.
Figures


Similar articles
-
Phosphorylation of the termini of Cauliflower mosaic virus precapsid protein is important for productive infection.Mol Plant Microbe Interact. 2007 Jun;20(6):648-58. doi: 10.1094/MPMI-20-6-0648. Mol Plant Microbe Interact. 2007. PMID: 17555273
-
Nuclear targeting of the cauliflower mosaic virus coat protein.J Virol. 1999 Jan;73(1):553-60. doi: 10.1128/JVI.73.1.553-560.1999. J Virol. 1999. PMID: 9847360 Free PMC article.
-
The cauliflower mosaic virus capsid protein: assembly and nucleic acid binding in vitro.Virus Genes. 1998;17(2):139-50. doi: 10.1023/a:1008064623335. Virus Genes. 1998. PMID: 9857987
-
Degradation signals within both terminal domains of the cauliflower mosaic virus capsid protein precursor.Plant J. 2001 Aug;27(4):335-43. doi: 10.1046/j.1365-313x.2001.01093.x. Plant J. 2001. PMID: 11532179
-
Plant pararetroviruses: interactions of cauliflower mosaic virus with plants and insects.Curr Opin Virol. 2013 Dec;3(6):629-38. doi: 10.1016/j.coviro.2013.08.014. Epub 2013 Sep 25. Curr Opin Virol. 2013. PMID: 24075119 Review.
Cited by
-
Phosphorylation of Beet black scorch virus coat protein by PKA is required for assembly and stability of virus particles.Sci Rep. 2015 Jun 25;5:11585. doi: 10.1038/srep11585. Sci Rep. 2015. PMID: 26108567 Free PMC article.
-
Phosphorylation of TGB1 by protein kinase CK2 promotes barley stripe mosaic virus movement in monocots and dicots.J Exp Bot. 2015 Aug;66(15):4733-47. doi: 10.1093/jxb/erv237. Epub 2015 May 21. J Exp Bot. 2015. PMID: 25998907 Free PMC article.
-
The N-Terminal Region of Cucumber Mosaic Virus 2a Protein Is Involved in the Systemic Infection in Brassica juncea.Plants (Basel). 2024 Mar 31;13(7):1001. doi: 10.3390/plants13071001. Plants (Basel). 2024. PMID: 38611534 Free PMC article.
-
Phosphorylation of plant virus proteins: Analysis methods and biological functions.Front Microbiol. 2022 Jul 26;13:935735. doi: 10.3389/fmicb.2022.935735. eCollection 2022. Front Microbiol. 2022. PMID: 35958157 Free PMC article. Review.
-
Phosphorylation of bamboo mosaic virus satellite RNA (satBaMV)-encoded protein P20 downregulates the formation of satBaMV-P20 ribonucleoprotein complex.Nucleic Acids Res. 2012 Jan;40(2):638-49. doi: 10.1093/nar/gkr705. Epub 2011 Sep 28. Nucleic Acids Res. 2012. PMID: 21965537 Free PMC article.
References
-
- Chapdelaine, Y., and T. Hohn. 1998. The cauliflower mosaic virus capsid protein: assembly and nucleic acid binding in vitro. Virus Genes 17:139-150. - PubMed
-
- Coffin, J. M. 1996. The scoop on HIV mutations. J. Int. Assoc. Physicians AIDS Care 2:45. - PubMed
-
- DuPlessis, D. H., and P. Smith. 1981. Glycosylation of the cauliflower mosaic virus capsid polypeptide. Virology 109:403-408. - PubMed
-
- Fütterer, J., and T. Hohn. 1987. Involvement of nucleocapsids in reverse transcription—a general phenomenon? Trends Biochem. Sci. 12:92-95.
-
- Gallay, P., S. Swingler, C. Aiken, and D. Trono. 1995. HIV-1 infection of non-dividing cells: C-terminal phosphorylation of the viral matrix protein is a key regulator. Cell 80:379-388. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

