Quantitative noise analysis for gene expression microarray experiments
- PMID: 12388780
- PMCID: PMC137831
- DOI: 10.1073/pnas.222164199
Quantitative noise analysis for gene expression microarray experiments
Abstract
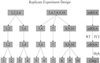
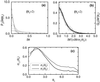
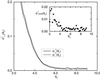
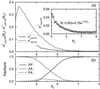
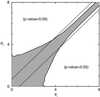
A major challenge in DNA microarray analysis is to effectively dissociate actual gene expression values from experimental noise. We report here a detailed noise analysis for oligonuleotide-based microarray experiments involving reverse transcription, generation of labeled cRNA (target) through in vitro transcription, and hybridization of the target to the probe immobilized on the substrate. By designing sets of replicate experiments that bifurcate at different steps of the assay, we are able to separate the noise caused by sample preparation and the hybridization processes. We quantitatively characterize the strength of these different sources of noise and their respective dependence on the gene expression level. We find that the sample preparation noise is small, implying that the amplification process during the sample preparation is relatively accurate. The hybridization noise is found to have very strong dependence on the expression level, with different characteristics for the low and high expression values. The hybridization noise characteristics at the high expression regime are mostly Poisson-like, whereas its characteristics for the small expression levels are more complex, probably due to cross-hybridization. A method to evaluate the significance of gene expression fold changes based on noise characteristics is proposed.
Figures
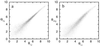

References
-
- Lockhart D. J. & Winzeler, E. A. (2000) Nature 405, 827-836. - PubMed
-
- Brown P. O. & Botstein, D. (1999) Nat. Genet. 21,Suppl., 33-37. - PubMed
-
- Novak J. P., Sladek, R. & Hudson, T. J. (2002) Genomics 79, 104-113. - PubMed
-
- Lockhart D. J., Dong, H., Byrne, M. C., Follettie, M. T., Gallo, M. V., Chee, M. S., Mittmann, M., Wang, C., Kobayashi, M., Hortan, H. & Brown, E. L. (1996) Nat. Biotechnol. 14, 1675-1680. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

