Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro
- PMID: 12391159
- PMCID: PMC134738
- DOI: 10.1128/MCB.22.22.7919-7928.2002
Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro
Abstract
Polycomb group (PcG) proteins are responsible for stable repression of homeotic gene expression during Drosophila melanogaster development. They are thought to stabilize chromatin structure to prevent transcription, though how they do this is unknown. We have established an in vitro system in which the PcG complex PRC1 and a recombinant PRC1 core complex (PCC) containing only PcG proteins are able to repress transcription by both RNA polymerase II and by T7 RNA polymerase. We find that assembly of the template into nucleosomes enhances repression by PRC1 and PCC. The subunit Psc is able to inhibit transcription on its own. PRC1- and PCC-repressed templates remain accessible to Gal4-VP16 binding, and incubation of the template with HeLa nuclear extract before the addition of PCC eliminates PCC repression. These results suggest that PcG proteins do not merely prohibit all transcription machinery from binding the template but instead likely inhibit specific steps in the transcription reaction.
Figures


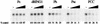


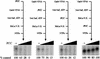
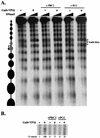
Similar articles
-
Reconstitution of a functional core polycomb repressive complex.Mol Cell. 2001 Sep;8(3):545-56. doi: 10.1016/s1097-2765(01)00316-1. Mol Cell. 2001. PMID: 11583617
-
Propagation of silencing; recruitment and repression of naive chromatin in trans by polycomb repressed chromatin.Mol Cell. 2004 Feb 13;13(3):415-25. doi: 10.1016/s1097-2765(04)00006-1. Mol Cell. 2004. PMID: 14967148
-
The role of the histone H2A ubiquitinase Sce in Polycomb repression.Development. 2012 Jan;139(1):117-27. doi: 10.1242/dev.074450. Epub 2011 Nov 17. Development. 2012. PMID: 22096074 Free PMC article.
-
Polycomb response elements and targeting of Polycomb group proteins in Drosophila.Curr Opin Genet Dev. 2006 Oct;16(5):476-84. doi: 10.1016/j.gde.2006.08.005. Epub 2006 Aug 17. Curr Opin Genet Dev. 2006. PMID: 16914306 Review.
-
Gene repression by Polycomb group protein complexes: a distinct complex for every occasion?Curr Opin Genet Dev. 2003 Oct;13(5):448-54. doi: 10.1016/s0959-437x(03)00108-4. Curr Opin Genet Dev. 2003. PMID: 14550408 Review.
Cited by
-
Mammalian polycomb-like Pcl2/Mtf2 is a novel regulatory component of PRC2 that can differentially modulate polycomb activity both at the Hox gene cluster and at Cdkn2a genes.Mol Cell Biol. 2011 Jan;31(2):351-64. doi: 10.1128/MCB.00259-10. Epub 2010 Nov 8. Mol Cell Biol. 2011. PMID: 21059868 Free PMC article.
-
Spreading of a corepressor linked to action of long-range repressor hairy.Mol Cell Biol. 2008 Apr;28(8):2792-802. doi: 10.1128/MCB.01203-07. Epub 2008 Feb 19. Mol Cell Biol. 2008. PMID: 18285466 Free PMC article.
-
Normal Patterns of Histone H3K27 Methylation Require the Histone Variant H2A.Z in Neurospora crassa.Genetics. 2020 Sep;216(1):51-66. doi: 10.1534/genetics.120.303442. Epub 2020 Jul 10. Genetics. 2020. PMID: 32651262 Free PMC article.
-
Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins.Proc Natl Acad Sci U S A. 2007 Oct 16;104(42):16615-20. doi: 10.1073/pnas.0701538104. Epub 2007 Oct 5. Proc Natl Acad Sci U S A. 2007. PMID: 17921257 Free PMC article.
-
Adaptive evolution of SCML1 in primates, a gene involved in male reproduction.BMC Evol Biol. 2008 Jul 5;8:192. doi: 10.1186/1471-2148-8-192. BMC Evol Biol. 2008. PMID: 18601738 Free PMC article.
References
-
- Akhtar, A., D. Zink, and P. B. Becker. 2000. Chromodomains are protein-RNA interaction modules. Nature 407:405-409. - PubMed
-
- Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1-13. - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
-
- Beuchle, D., G. Struhl, and J. Muller. 2001. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128:993-1004. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
