High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction
- PMID: 12391171
- PMCID: PMC134739
- DOI: 10.1128/MCB.22.22.8056-8066.2002
High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction
Abstract
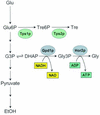
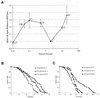
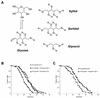
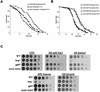
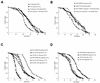
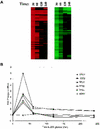
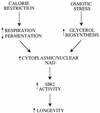
Calorie restriction (CR) extends life span in many different organisms, including mammals. We describe here a novel pathway that extends the life span of Saccharomyces cerevisiae mother cells but does not involve a reduction in caloric content of the media, i.e., there is growth of yeast cells in the presence of a high concentration of external osmolytes. Like CR, this longevity-promoting response to high osmolarity requires SIR2, suggesting a common mechanism of life span regulation. Genetic and microarray analysis indicates that high osmolarity extends the life span by activating Hog1p, leading to an increase in the biosynthesis of glycerol from glycolytic intermediates. This metabolic shift likely increases NAD levels, thereby activating Sir2p and promoting longevity.
Figures

References
-
- Albertyn, J., S. Hohmann, J. M. Thevelein, and B. A. Prior. 1994. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14:4135-4144. - PMC - PubMed
-
- Alepuz, P. M., A. Jovanovic, V. Reiser, and G. Ammerer. 2001. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell 7:767-777. - PubMed
-
- Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, H. Cohen, S. S. Lin, J. K. Manchester, J. I. Gordon, and D. A. Sinclair. 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277:18881-18890. - PubMed
-
- Bell, M., R. Capone, I. Pashtan, A. Levitzki, and D. Engelberg. 2001. Isolation of hyperactive mutants of the MAPK p38/Hog1 that are independent of MAPK kinase activation. J. Biol. Chem. 276:25351-25358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
