Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain
- PMID: 12391316
- PMCID: PMC137845
- DOI: 10.1073/pnas.172527399
Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain
Abstract
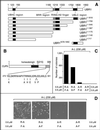
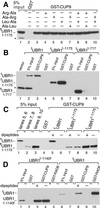
Protein degradation by the ubiquitin (Ub) system controls the concentrations of many regulatory proteins. The degradation signals (degrons) of these proteins are recognized by the system's Ub ligases (complexes of E2 and E3 enzymes). Two substrate-binding sites of UBR1, the E3 of the N-end rule pathway in the yeast Saccharomyces cerevisiae, recognize basic (type 1) and bulky hydrophobic (type 2) N-terminal residues of proteins or short peptides. A third substrate-binding site of UBR1 targets CUP9, a transcriptional repressor of the peptide transporter PTR2, through an internal (non-N-terminal) degron of CUP9. Previous work demonstrated that dipeptides with destabilizing N-terminal residues allosterically activate UBR1, leading to accelerated in vivo degradation of CUP9 and the induction of PTR2 expression. Through this positive feedback, S. cerevisiae can sense the presence of extracellular peptides and react by accelerating their uptake. Here, we show that dipeptides with destabilizing N-terminal residues cause dissociation of the C-terminal autoinhibitory domain of UBR1 from its N-terminal region that contains all three substrate-binding sites. This dissociation, which allows the interaction between UBR1 and CUP9, is strongly increased only if both type 1- and type 2-binding sites of UBR1 are occupied by dipeptides. An aspect of autoinhibition characteristic of yeast UBR1 also was observed with mammalian (mouse) UBR1. The discovery of autoinhibition in Ub ligases of the UBR family indicates that this regulatory mechanism may also control the activity of other Ub ligases.
Figures



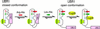
References
-
- Hershko A., Ciechanover, A. & Varshavsky, A. (2000) Nat. Med. 10, 1073-1081. - PubMed
-
- Weissman A. M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 169-178. - PubMed
-
- Pickart C. (2001) Annu. Rev. Biochem. 70, 503-533. - PubMed
-
- Conaway R. C., Brower, C. S. & Conaway, J. W. (2002) Science 296, 1254-1258. - PubMed
-
- Zwickl P., Baumeister, W. & Steven, A. (2000) Curr. Opin. Struct. Biol. 10, 242-250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

