Transcriptome analysis during cell division in plants
- PMID: 12393816
- PMCID: PMC137503
- DOI: 10.1073/pnas.222561199
Transcriptome analysis during cell division in plants
Abstract
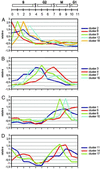
Using synchronized tobacco Bright Yellow-2 cells and cDNA-amplified fragment length polymorphism-based genomewide expression analysis, we built a comprehensive collection of plant cell cycle-modulated genes. Approximately 1,340 periodically expressed genes were identified, including known cell cycle control genes as well as numerous unique candidate regulatory genes. A number of plant-specific genes were found to be cell cycle modulated. Other transcript tags were derived from unknown plant genes showing homology to cell cycle-regulatory genes of other organisms. Many of the genes encode novel or uncharacterized proteins, indicating that several processes underlying cell division are still largely unknown.
Figures



References
-
- Cho R. J., Huang, M., Campbell, M. J., Dong, H., Steinmetz, L., Sapinoso, L., Hampton, G., Elledge, S. J., Davis, R. W. & Lockhart, D. J. (2001) Nat. Genet. 27, 48-54. - PubMed
-
- Stals H. & Inzé, D. (2001) Trends Plant Sci. 6, 359-364. - PubMed
-
- den Boer B. G. W. & Murray, J. A. H. (2000) Trends Cell Biol. 10, 245-250. - PubMed
-
- Smith L. G. (1999) Curr. Opin. Plant Biol. 2, 447-453. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

