Metabolic specialization associated with phenotypic switching in Candidaalbicans
- PMID: 12397174
- PMCID: PMC137518
- DOI: 10.1073/pnas.232566499
Metabolic specialization associated with phenotypic switching in Candidaalbicans
Abstract
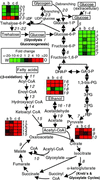
Phase and antigenic variation are mechanisms used by microbial pathogens to stochastically change their cell surface composition. A related property, referred to as phenotypic switching, has been described for some pathogenic fungi. This phenomenon is best studied in Candida albicans, where switch phenotypes vary in morphology, physiology, and pathogenicity in experimental models. In this study, we report an application of a custom Affymetrix GeneChip representative of the entire C. albicans genome and assay the global expression profiles of white and opaque switch phenotypes of the WO-1 strain. Of 13,025 probe sets examined, 373 ORFs demonstrated a greater than twofold difference in expression level between switch phenotypes. Among these, 221 were expressed at a level higher in opaque cells than in white cells; conversely, 152 were more highly expressed in white cells. Affected genes represent functions as diverse as metabolism, adhesion, cell surface composition, stress response, signaling, mating type, and virulence. Approximately one-third of the differences between cell types are related to metabolic pathways, opaque cells expressing a transcriptional profile consistent with oxidative metabolism and white cells expressing a fermentative one. This bias was obtained regardless of carbon source, suggesting a connection between phenotypic switching and metabolic flexibility, where metabolic specialization of switch phenotypes enhances selection in relation to the nutrients available at different anatomical sites. These results extend our understanding of strategies used in microbial phase variation and pathogenesis and further characterize the unanticipated diversity of genes expressed in phenotypic switching.
Figures



Similar articles
-
Analysis of phase-specific gene expression at the single-cell level in the white-opaque switching system of Candida albicans.J Bacteriol. 2001 Jun;183(12):3761-9. doi: 10.1128/JB.183.12.3761-3769.2001. J Bacteriol. 2001. PMID: 11371541 Free PMC article.
-
The white-phase-specific gene WH11 is not required for white-opaque switching in Candida albicans.Mol Genet Genomics. 2004 Aug;272(1):88-97. doi: 10.1007/s00438-004-1037-1. Epub 2004 Jul 13. Mol Genet Genomics. 2004. PMID: 15249973
-
Candida albicans MTLalpha tup1Delta mutants can reversibly switch to mating-competent, filamentous growth forms.Mol Microbiol. 2005 Dec;58(5):1288-302. doi: 10.1111/j.1365-2958.2005.04898.x. Mol Microbiol. 2005. PMID: 16313617
-
Mating-type locus homozygosis, phenotypic switching and mating: a unique sequence of dependencies in Candida albicans.Bioessays. 2004 Jan;26(1):10-20. doi: 10.1002/bies.10379. Bioessays. 2004. PMID: 14696036 Review.
-
High-frequency phenotypic switching in Candida albicans.Trends Genet. 1993 Feb;9(2):61-5. doi: 10.1016/0168-9525(93)90189-O. Trends Genet. 1993. PMID: 8456504 Review.
Cited by
-
Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism.Nat Genet. 2013 Sep;45(9):1088-91. doi: 10.1038/ng.2710. Epub 2013 Jul 28. Nat Genet. 2013. PMID: 23892606 Free PMC article.
-
Chromatin Structure and Drug Resistance in Candida spp.J Fungi (Basel). 2020 Jul 30;6(3):121. doi: 10.3390/jof6030121. J Fungi (Basel). 2020. PMID: 32751495 Free PMC article. Review.
-
Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop.Proc Natl Acad Sci U S A. 2006 Aug 22;103(34):12807-12. doi: 10.1073/pnas.0605138103. Epub 2006 Aug 9. Proc Natl Acad Sci U S A. 2006. PMID: 16899543 Free PMC article.
-
Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans.Proc Natl Acad Sci U S A. 2006 Aug 22;103(34):12813-8. doi: 10.1073/pnas.0605270103. Epub 2006 Aug 11. Proc Natl Acad Sci U S A. 2006. PMID: 16905649 Free PMC article.
-
Candida albicans can foster gut dysbiosis and systemic inflammation during HIV infection.Gut Microbes. 2023 Jan-Dec;15(1):2167171. doi: 10.1080/19490976.2023.2167171. Gut Microbes. 2023. PMID: 36722096 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

