Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis
- PMID: 12399499
- PMCID: PMC151947
- DOI: 10.1128/JB.184.22.6289-6300.2002
Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis
Abstract
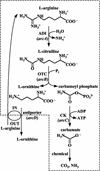
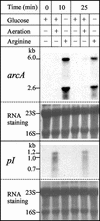
Although Enteroccus faecalis is the paradigm for biochemical studies on the arginine deiminase (ADI) pathway of fermentative arginine catabolism, little genetic information exists on this pathway in this organism. We fill this important gap by characterizing, in an 8,228-bp region cloned from a lambdagt11 genomic library of E. faecalis, a five-gene cluster forming a transcriptional unit (revealed by Northern blots and primer extension in E. faecalis) that corresponds to the ADI operon. Four additional genes in the opposite DNA strand and one in the same DNA strand are also identified. Studies on the protein products, including heterologous expression and/or sequence comparisons, allow us to ascertain or propose functions for all but 1 of the 10 genes. The ADI operon genes, arcABCRD, encode, respectively, ADI, ornithine transcarbamylase, carbamate kinase, a putative Crp/Fnr-type regulator (ArcR), and a putative ornithine-arginine antiporter (ArcD). Arginine induces the expression of arcABCRD, most likely by means of two homologous ArgR/AhrC-type regulators encoded by two genes, argR1 and argR2, that precede arcABCRD in each DNA strand and that are transcribed monocistronically, their transcription being influenced differentially by glucose and arginine. Potential ArgR1/ArgR2 (double and single) binding sequences are found in the promoter regions of arcA and of argR1/argR2 themselves. In addition, putative binding sequences for ArcR and for CcpA are found, respectively, in the argR1/argR2 and arcA promoter regions. Of the three other genes identified, two form a transcriptional unit and encode a putative metal-sensitive transcriptional regulator (ArsR) and a cysteine protease.
Figures





References
-
- Abdelal, A. T. 1979. Arginine catabolism by microorganisms. Annu. Rev. Microbiol. 33:139-168. - PubMed
-
- Allen, A. G., and R. N. Perham. 1991. Two lipoyl domains in the dihydrolipoamide acetyltransferase chain of the pyruvate dehydrogenase multienzyme complex of Streptococcus faecalis. FEBS Lett. 287:206-210. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
-
- Baur, H., E. Luethi, V. Stalon, A. Mercenier, and D. Haas. 1989. Sequence analysis and expression of the arginine-deiminase and carbamate-kinase genes of Pseudomonas aeruginosa. Eur. J. Biochem. 179:53-60. - PubMed
-
- Baur, H., V. Stalon, P. Falmagne, E. Luethi, and D. Haas. 1987. Primary and quaternary structure of the catabolic ornithine carbamoyltransferase from Pseudomonas aeruginosa. Eur. J. Biochem. 166:111-117. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

