SeqHound: biological sequence and structure database as a platform for bioinformatics research
- PMID: 12401134
- PMCID: PMC138791
- DOI: 10.1186/1471-2105-3-32
SeqHound: biological sequence and structure database as a platform for bioinformatics research
Abstract
Background: SeqHound has been developed as an integrated biological sequence, taxonomy, annotation and 3-D structure database system. It provides a high-performance server platform for bioinformatics research in a locally-hosted environment.
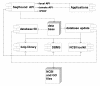
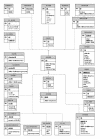
Results: SeqHound is based on the National Center for Biotechnology Information data model and programming tools. It offers daily updated contents of all Entrez sequence databases in addition to 3-D structural data and information about sequence redundancies, sequence neighbours, taxonomy, complete genomes, functional annotation including Gene Ontology terms and literature links to PubMed. SeqHound is accessible via a web server through a Perl, C or C++ remote API or an optimized local API. It provides functionality necessary to retrieve specialized subsets of sequences, structures and structural domains. Sequences may be retrieved in FASTA, GenBank, ASN.1 and XML formats. Structures are available in ASN.1, XML and PDB formats. Emphasis has been placed on complete genomes, taxonomy, domain and functional annotation as well as 3-D structural functionality in the API, while fielded text indexing functionality remains under development. SeqHound also offers a streamlined WWW interface for simple web-user queries.
Conclusions: The system has proven useful in several published bioinformatics projects such as the BIND database and offers a cost-effective infrastructure for research. SeqHound will continue to develop and be provided as a service of the Blueprint Initiative at the Samuel Lunenfeld Research Institute. The source code and examples are available under the terms of the GNU public license at the Sourceforge site http://sourceforge.net/projects/slritools/ in the SLRI Toolkit.
Figures


References
-
- Schuler GD, Epstein JA, Ohkawa H, Kans JA. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–162. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

