Visualization of replication initiation and elongation in Drosophila
- PMID: 12403810
- PMCID: PMC2173051
- DOI: 10.1083/jcb.200207046
Visualization of replication initiation and elongation in Drosophila
Abstract
Chorion gene amplification in the ovaries of Drosophila melanogaster is a powerful system for the study of metazoan DNA replication in vivo. Using a combination of high-resolution confocal and deconvolution microscopy and quantitative realtime PCR, we found that initiation and elongation occur during separate developmental stages, thus permitting analysis of these two phases of replication in vivo. Bromodeoxyuridine, origin recognition complex, and the elongation factors minichromosome maintenance proteins (MCM)2-7 and proliferating cell nuclear antigen were precisely localized, and the DNA copy number along the third chromosome chorion amplicon was quantified during multiple developmental stages. These studies revealed that initiation takes place during stages 10B and 11 of egg chamber development, whereas only elongation of existing replication forks occurs during egg chamber stages 12 and 13. The ability to distinguish initiation from elongation makes this an outstanding model to decipher the roles of various replication factors during metazoan DNA replication. We utilized this system to demonstrate that the pre-replication complex component, double-parked protein/cell division cycle 10-dependent transcript 1, is not only necessary for proper MCM2-7 localization, but, unexpectedly, is present during elongation.
Figures

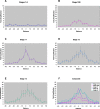
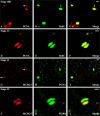
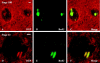
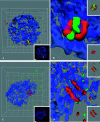
Similar articles
-
Localisation of the DmCdc45 DNA replication factor in the mitotic cycle and during chorion gene amplification.Nucleic Acids Res. 2000 Oct 15;28(20):3897-903. doi: 10.1093/nar/28.20.3897. Nucleic Acids Res. 2000. PMID: 11024168 Free PMC article.
-
Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication.Mol Biol Cell. 2002 Feb;13(2):607-20. doi: 10.1091/mbc.01-08-0400. Mol Biol Cell. 2002. PMID: 11854416 Free PMC article.
-
Sequence requirements for function of the Drosophila chorion gene locus ACE3 replicator and ori-beta origin elements.Development. 2004 May;131(9):2089-99. doi: 10.1242/dev.01064. Development. 2004. PMID: 15105371
-
MCM proteins in DNA replication.Annu Rev Biochem. 1999;68:649-86. doi: 10.1146/annurev.biochem.68.1.649. Annu Rev Biochem. 1999. PMID: 10872463 Review.
-
Regulation of chromosome replication.Annu Rev Biochem. 2000;69:829-80. doi: 10.1146/annurev.biochem.69.1.829. Annu Rev Biochem. 2000. PMID: 10966477 Review.
Cited by
-
Replication fork progression during re-replication requires the DNA damage checkpoint and double-strand break repair.Curr Biol. 2015 Jun 15;25(12):1654-60. doi: 10.1016/j.cub.2015.04.058. Epub 2015 Jun 4. Curr Biol. 2015. PMID: 26051888 Free PMC article.
-
Finishing the egg.Genetics. 2024 Jan 3;226(1):iyad183. doi: 10.1093/genetics/iyad183. Genetics. 2024. PMID: 38000906 Free PMC article.
-
Quantitative real-time PCR with SYBR Green detection to assess gene duplication in insects: study of gene dosage in Drosophila melanogaster (Diptera) and in Ostrinia nubilalis (Lepidoptera).BMC Res Notes. 2011 Mar 28;4:84. doi: 10.1186/1756-0500-4-84. BMC Res Notes. 2011. PMID: 21443764 Free PMC article.
-
The role of transcription in the activation of a Drosophila amplification origin.G3 (Bethesda). 2014 Oct 14;4(12):2403-8. doi: 10.1534/g3.114.014050. G3 (Bethesda). 2014. PMID: 25320071 Free PMC article.
-
Adult neurogenesis: a common strategy across diverse species.J Comp Neurol. 2007 Jan 20;500(3):574-84. doi: 10.1002/cne.21187. J Comp Neurol. 2007. PMID: 17120293 Free PMC article.
References
-
- Aparicio, O.M., D.M. Weinstein, and S.P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 91:59–69. - PubMed
-
- Ashburner, M. 1989. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 434 pp.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

