Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice
- PMID: 12403827
- PMCID: PMC137533
- DOI: 10.1073/pnas.222425399
Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice
Abstract
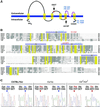
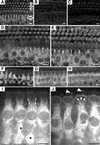
Deafness in spontaneously occurring mouse mutants is often associated with defects in cochlea sensory hair cells, opening an avenue to systematically identify genes critical for hair cell structure and function. The classical semidominant mouse mutant varitint-waddler (Va) exhibits early-onset hearing loss, vestibular defects, pigmentation abnormalities, and perinatal lethality. A second allele, Va(J), which arose in a cross segregating for Va, shows a less severe phenotype. By using a positional cloning strategy, we identify two additional members of the mucolipin gene family (Mcoln2 and Mcoln3) in the 350-kb Va(J) minimal interval and provide evidence for Mcoln3 as the gene mutated in varitint-waddler. Mcoln3 encodes a putative six-transmembrane-domain protein with sequence and motif similarities to the family of nonselective transient-receptor-potential (TRP) ion channels. In the Va allele an Ala419Pro substitution occurs in the fifth transmembrane domain of Mcoln3, and in Va(J), a second sequence alteration (Ile362Thr) occurring in cis partially rescues the Va allele. Mcoln3 localizes to cytoplasmic compartments of hair cells and plasma membrane of stereocilia. Hair cell defects are apparent by embryonic day 17.5, assigning Mcoln3 an essential role during early hair cell maturation. Our data suggest that Mcoln3 is involved in ion homeostasis and acts cell-autonomously. Hence, we identify a molecular link between hair cell physiology and melanocyte function. Last, MCOLN2 and MCOLN3 are candidate genes for hereditary and/or sporadic forms of neurosensory disorders in humans.
Figures


Comment in
-
Varitint-waddler: a double whammy for hearing.Proc Natl Acad Sci U S A. 2002 Nov 12;99(23):14613-5. doi: 10.1073/pnas.232585699. Epub 2002 Nov 4. Proc Natl Acad Sci U S A. 2002. PMID: 12417743 Free PMC article. No abstract available.
References
-
- Steel K. P. & Kros, C. J. (2001) Nat. Genet. 27 143-149. - PubMed
-
- Geissler E. N., Ryan, M. A. & Housman, D. E. (1988) Cell 55 185-192. - PubMed
-
- Chabot B., Stephenson, D. A., Chapman, V. M., Besmer, P. & Bernstein, A. (1988) Nature 335 88-89. - PubMed
-
- Matsui Y., Zsebo, K. M. & Hogan, B. L. (1990) Nature 347 667-669. - PubMed
-
- Huang E., Nocka, K., Beier, D. R., Chu, T. Y., Buck, J., Lahm, H. W., Wellner, D., Leder, P. & Besmer, P. (1990) Cell 63 225-233. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

