A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco
- PMID: 12403829
- PMCID: PMC137572
- DOI: 10.1073/pnas.232434999
A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco
Abstract
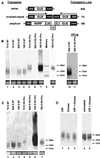
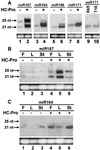
Two major classes of small noncoding RNAs have emerged as important regulators of gene expression in eukaryotes, the short interfering RNAs (siRNAs) associated with RNA silencing and endogenous micro-RNAs (miRNAs) implicated in regulation of gene expression. Helper component-proteinase (HC-Pro) is a viral protein that blocks RNA silencing in plants. Here we examine the effect of HC-Pro on the accumulation of siRNAs and endogenous miRNAs. siRNAs were analyzed in transgenic tobacco plants silenced in response to three different classes of transgenes: sense-transgenes, inverted-repeat transgenes, and amplicon-transgenes. HC-Pro suppressed silencing in each line, blocking accumulation of the associated siRNAs and allowing accumulation of transcripts from the previously silenced loci. HC-Pro-suppression of silencing in the inverted-repeat- and amplicon-transgenic lines was accompanied by the apparent accumulation of long double-stranded RNAs and proportional amounts of small RNAs that are larger than the siRNAs that accumulate during silencing. Analysis of these results suggests that HC-Pro interferes with silencing either by inhibiting siRNA processing from double-stranded RNA precursors or by destabilizing siRNAs. In contrast to siRNAs, the accumulation of endogenous miRNAs was greatly enhanced in all of the HC-Pro-expressing lines. Thus, our results demonstrate that accumulation of siRNAs and miRNAs in plants can be differentially regulated by a viral protein. The fact that HC-Pro affects the miRNA pathway raises the possibility that this pathway is targeted by plant viruses as a means to control gene expression in the host.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

