Structural analysis of insulin minisatellite alleles reveals unusually large differences in diversity between Africans and non-Africans
- PMID: 12404181
- PMCID: PMC378563
- DOI: 10.1086/344513
Structural analysis of insulin minisatellite alleles reveals unusually large differences in diversity between Africans and non-Africans
Abstract
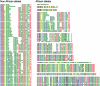
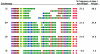
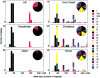
The insulin minisatellite (INS VNTR) associates with susceptibility to a variety of diseases. We have developed a high-resolution system for analyzing variant repeat distributions applicable to all known minisatellite alleles, irrespective of size, which allows lineages of related alleles to be identified. This system has previously revealed extremely low structural diversity in the minisatellite among northern Europeans from the United Kingdom, with all alleles belonging to one of only three highly diverged lineages called "I," "IIIA," and "IIIB." To explore the origins of this remarkably limited lineage diversity, we have characterized an additional 780 alleles from three non-African and three African populations. In total, 22 highly diverged lineages were identified, with structural intermediates absent from extant populations, suggesting a bottleneck within the ancestry of all humans. The difference between levels of diversity in Africans and non-Africans is unusually large, with all 22 lineages identified in Africa compared with only three lineages seen not only in the United Kingdom but also in the other non-African populations. We also find evidence for overrepresentation of lineage I chromosomes in non-Africans. These data are consistent with a common out-of-Africa origin and an unusually tight bottleneck within the ancestry of all non-African populations, possibly combined with differential and positive selection for lineage I alleles in non-Africans. The important implications of these data for future disease-association studies are discussed.
Figures


References
Electronic-Database Information
-
- Variant Repeat Mapping at the Insulin Minisatellite, http://www.leicester.ac.uk/genetics/ajj/insulin/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for INS VNTR [MIM 125852])
References
-
- Armour JA, Anttinen T, May CA, Vega EE, Sajantila A, Kidd JR, Kidd KK, Bertranpetit J, Paabo S, Jeffreys AJ (1996) Minisatellite diversity supports a recent African origin for modern humans. Nat Genet 13:154–160 - PubMed
-
- Bennett ST, Lucassen AM, Gough SC, Powell EE, Undlien DE, Pritchard LE, Merriman ME, Kawaguchi Y, Dronsfield MJ, Pociot F, Nerup J, Bouzekri N, Cambon-Thomsen A, Ronningen KS, Barnett AH, Bain SC, Todd JA (1995) Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet 9:284–292 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical

