Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers
- PMID: 12406765
- PMCID: PMC129938
- DOI: 10.1128/AEM.68.11.5685-5692.2002
Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers
Erratum in
- Appl Environ Microbiol. 2003 May;69(5):3027
Abstract
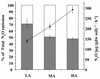
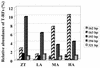
The effect of ammonium addition (6.5, 58, and 395 microg of NH4+-N g [dry weight] of soil(-1)) on soil microbial communities was explored. For medium and high ammonium concentrations, increased N2O release rates and a shift toward a higher contribution of nitrification to N2O release occurred after incubation for 5 days at 4 degrees C. Communities of ammonia oxidizers were assayed after 4 weeks of incubation by denaturant gradient gel electrophoresis (DGGE) of the amoA gene coding for the small subunit of ammonia monooxygenase. The DGGE fingerprints were invariably the same whether the soil was untreated or incubated with low, medium, or high ammonium concentrations. Phylogenetic analysis of cloned PCR products from excised DGGE bands detected amoA sequences which probably belonged to Nitrosospira 16S rRNA clusters 3 and 4. Additional clones clustered with Nitrosospira sp. strains Ka3 and Ka4 and within an amoA cluster from unknown species. A Nitrosomonas-like amoA gene was detected in only one clone. In agreement with the amoA results, community profiles of total bacteria analyzed by terminal restriction fragment length polymorphism (T-RFLP) showed only minor differences. However, a community shift occurred for denitrifier populations based on T-RFLP analysis of nirK genes encoding copper-containing nitrite reductase with incubation at medium and high ammonia concentrations. Major terminal restriction fragments observed in environmental samples were further described by correspondence to cloned nirK genes from the same soil. Phylogenetic analysis grouped these clones into clusters of soil nirK genes. However, some clones were also closely related to genes from known denitrifiers. The shift in the denitrifier community was probably the consequence of the increased supply of oxidized nitrogen through nitrification. Nitrification activity increased upon addition of ammonium, but the community structure of ammonium oxidizers did not change.
Figures



References
-
- Aakra, A., J. B. Utaker, and I. F. Nes. 2001. Comparative phylogeny of the ammonia monooxygenase subunit A and 16S rRNA genes of ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 205:237-242. - PubMed
-
- Aakra, A., J. B. Utaker, A. Pommerening-Roser, H. P. Koops, and I. F. Nes. 2001. Detailed phylogeny of ammonia-oxidizing bacteria determined by rDNA sequences and DNA homology values. Int. J. Syst. E vol. Microbiol. 51:2021-2030. - PubMed
-
- Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

