Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton
- PMID: 12406771
- PMCID: PMC129917
- DOI: 10.1128/AEM.68.11.5728-5736.2002
Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton
Abstract
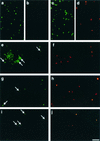
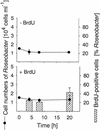
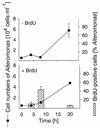
We describe a method for microscopic identification of DNA-synthesizing cells in bacterioplankton samples. After incubation with the halogenated thymidine analogue bromodeoxyuridine (BrdU), environmental bacteria were identified by fluorescence in situ hybridization (FISH) with horseradish peroxidase (HRP)-linked oligonucleotide probes. Tyramide signal amplification was used to preserve the FISH staining during the subsequent immunocytochemical detection of BrdU incorporation. DNA-synthesizing cells were visualized by means of an HRP-labeled antibody Fab fragment and a second tyramide signal amplification step. We applied our protocol to samples of prefiltered (pore size, 1.2 micro m) North Sea surface water collected during early autumn. After 4 h of incubation, BrdU incorporation was detected in 3% of all bacterial cells. Within 20 h the detectable DNA-synthesizing fraction increased to >14%. During this period, the cell numbers of members of the Roseobacter lineage remained constant, but the fraction of BrdU-incorporating Roseobacter sp. cells doubled, from 24 to 42%. In Alteromonas sp. high BrdU labeling rates after 4 to 8 h were followed by a 10-fold increase in abundance. Rapid BrdU incorporation was also observed in members of the SAR86 lineage. After 4 h of incubation, cells affiliated with this clade constituted 8% of the total bacteria but almost 50% of the visibly DNA-synthesizing bacterial fraction. Thus, this clade might be an important contributor to total bacterioplankton activity in coastal North Sea water during periods of low phytoplankton primary production. The small size and low ribosome content of SAR86 cells are probably not indications of inactivity or dormancy.
Figures


References
-
- Bak, P. M., and R. J. Panos. 1997. Protease antigen recovery decreases the specificity of bromodeoxyuridine detection in formalin-fixed tissue. J. Histochem. Cytochem. 45:1165-1170. - PubMed
-
- Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. - PubMed
-
- Binnie, C., and J. G. Coote. 1986. Inhibition of sporulation in Bacillus subtilis by bromodeoxyuridine and the effect on DNA replication. J. Gen. Microbiol. 132:493-502. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

