Negative autoregulation of BCL-6 is bypassed by genetic alterations in diffuse large B cell lymphomas
- PMID: 12407182
- PMCID: PMC137537
- DOI: 10.1073/pnas.232581199
Negative autoregulation of BCL-6 is bypassed by genetic alterations in diffuse large B cell lymphomas
Erratum in
- Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):17222.
Abstract
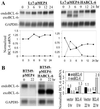
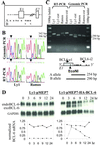
Thirty to forty percent of diffuse large B cell lymphomas (DLBCL) carry BCL-6 translocations that disrupt its 5' regulatory region. This same region is also subject to somatic hypermutations, although only a small fraction of these mutations have a detectable effect on transcription. Here, we show that transcription of the BCL-6 gene is negatively self-regulated in multiple cell types. This mechanism operates by means of the interaction of two BCL-6-binding sites within exon 1 of the gene and the BCL-6 protein itself, which is a potent transcription repressor. Because the DLBCL-associated "activating mutations" specifically target these exon 1 binding sites, and because the entire exon 1 is usually removed in the BCL-6-translocated tumors, this autoregulation is bypassed in 30-40% of all DLBCL cases. Our results not only demonstrate an important mechanism governing the expression of BCL-6, but also explain how BCL-6 is deregulated in a large number of DLBCL patients, providing a better understanding of BCL-6-related lymphomagenesis.
Figures




Similar articles
-
BCL-6 mRNA expression in higher grade transformation of follicle center lymphoma: correlation with somatic mutations in the 5' regulatory region of the BCL-6 gene.Leukemia. 2002 Sep;16(9):1857-62. doi: 10.1038/sj.leu.2402578. Leukemia. 2002. PMID: 12200704
-
Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma.Blood. 2003 Apr 15;101(8):2914-23. doi: 10.1182/blood-2002-11-3387. Epub 2002 Dec 19. Blood. 2003. PMID: 12515714
-
Follicle center lymphoma is associated with significantly elevated levels of BCL-6 expression among lymphoma subtypes, independent of chromosome 3q27 rearrangements.Leukemia. 2002 Nov;16(11):2318-25. doi: 10.1038/sj.leu.2402657. Leukemia. 2002. PMID: 12399978
-
The proto-oncogene BCL-6 in normal and malignant B cell development.Hematol Oncol. 2002 Dec;20(4):155-66. doi: 10.1002/hon.689. Hematol Oncol. 2002. PMID: 12469325 Review.
-
BCL-6 in diffuse large-cell lymphomas.Important Adv Oncol. 1996:139-48. Important Adv Oncol. 1996. PMID: 8791133 Review. No abstract available.
Cited by
-
POZ-, AT-hook-, and zinc finger-containing protein (PATZ) interacts with human oncogene B cell lymphoma 6 (BCL6) and is required for its negative autoregulation.J Biol Chem. 2012 May 25;287(22):18308-17. doi: 10.1074/jbc.M112.346270. Epub 2012 Apr 9. J Biol Chem. 2012. Retraction in: J Biol Chem. 2017 Mar 31;292(13):5609. doi: 10.1074/jbc.A117.346270. PMID: 22493480 Free PMC article. Retracted.
-
Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas.Blood. 2008 Feb 1;111(3):1515-23. doi: 10.1182/blood-2007-04-087734. Epub 2007 Oct 19. Blood. 2008. PMID: 17951530 Free PMC article.
-
Common mechanisms for the regulation of B cell differentiation and transformation by the transcriptional repressor protein BCL-6.Immunol Res. 2007;37(3):177-86. doi: 10.1007/BF02697368. Immunol Res. 2007. PMID: 17873402 Review.
-
Mouse Models of Germinal Center Derived B-Cell Lymphomas.Front Immunol. 2021 Aug 12;12:710711. doi: 10.3389/fimmu.2021.710711. eCollection 2021. Front Immunol. 2021. PMID: 34456919 Free PMC article. Review.
-
Antibody diversification: mutational mechanisms and oncogenesis.Immunol Res. 2006;35(1-2):75-88. doi: 10.1385/IR:35:1:75. Immunol Res. 2006. PMID: 17003511 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

