Genetic characterization and sequence heterogeneity of a canadian isolate of Swine hepatitis E virus
- PMID: 12409369
- PMCID: PMC139705
- DOI: 10.1128/JCM.40.11.4021-4029.2002
Genetic characterization and sequence heterogeneity of a canadian isolate of Swine hepatitis E virus
Abstract
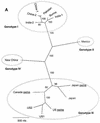
Swine hepatitis E virus (HEV) is a newly identified potentially zoonotic agent that is possibly transmitted to humans from pigs. Swine HEV is prevalent in pig populations and does not cause abnormal clinical symptoms in infected pigs, further implicating a likelihood of a risk of transmission to humans by normal contact. To date in North America, only one strain of swine HEV (strain US swine) has been fully sequenced. In the present study, we identified a swine HEV isolate from pigs in Canada, designated the Arkell strain, and determined the full length of the genomic sequence. The genome of Canadian strain Arkell consisted of 7,242 nucleotides, excluding the poly(A) tail of at least 15 A residues. The genome contained three open reading frames (ORFs), ORF1, ORF2, and ORF3, which had coding capacities for proteins of 1,708, 660, and 122 amino acids, respectively. Comparative analysis of the full-length genomic sequence indicated that the sequence of strain Arkell was distinct from those of all other known HEV isolates by 13 to 27% and shared the highest degrees of identity with human HEV isolates US-1 and US-2, HEV isolate US swine, and the human and swine HEV isolates recently isolated in Japan. On the basis of sequence similarities and phylogenetic analyses, HEV strain Arkell was grouped into genotype 3. The sequence of the Arkell swine HEV isolate differed from those of HEV isolate US swine and HEV isolate Japan swine by 13 and 14%, respectively. To date, two isolates of swine HEV (isolates Arkell and SK3 [D. Yoo et al., Clin. Diagn. Lab. Immunol. 8:1213-1219, 2001]) have been identified in Canadian pigs, and their sequences also differ from each other by 11.8%. Our studies indicate that, as with human HEV strains, swine HEV isolates exhibit extensive genetic heterogeneity.
Figures


Similar articles
-
Full-genome nucleotide sequence and analysis of a Chinese swine hepatitis E virus isolate of genotype 4 identified in the Guangxi Zhuang autonomous region: evidence of zoonotic risk from swine to human in South China.Liver Int. 2009 Sep;29(8):1230-40. doi: 10.1111/j.1478-3231.2009.02012.x. Epub 2009 Apr 15. Liver Int. 2009. PMID: 19490423
-
Full genome sequence and analysis of Indian swine hepatitis E virus isolate of genotype 4.Vet Microbiol. 2006 May 31;114(3-4):240-51. doi: 10.1016/j.vetmic.2005.12.007. Epub 2006 Feb 2. Vet Microbiol. 2006. PMID: 16457969
-
Analysis of the complete genome of indigenous swine hepatitis E virus isolated in Japan.Biochem Biophys Res Commun. 2001 Dec 21;289(5):929-36. doi: 10.1006/bbrc.2001.6088. Biochem Biophys Res Commun. 2001. PMID: 11741279
-
Recent advances in Hepatitis E virus.J Viral Hepat. 2010 Mar;17(3):153-61. doi: 10.1111/j.1365-2893.2009.01257.x. Epub 2009 Dec 21. J Viral Hepat. 2010. PMID: 20040046 Review.
-
Multifunctional ORF3 protein of hepatitis E virus.J Med Virol. 2024 Jun;96(6):e29691. doi: 10.1002/jmv.29691. J Med Virol. 2024. PMID: 38783788 Review.
Cited by
-
Phylogenetic analysis of two new complete genomes of the hepatitis E virus (HEV) genotype 3 from Thailand.Mol Biol Rep. 2020 Nov;47(11):8657-8668. doi: 10.1007/s11033-020-05908-3. Epub 2020 Oct 14. Mol Biol Rep. 2020. PMID: 33058031 Free PMC article.
-
Molecular biology and pathogenesis of hepatitis E virus.J Biosci. 2008 Nov;33(4):451-64. doi: 10.1007/s12038-008-0064-1. J Biosci. 2008. PMID: 19208971 Review.
-
Molecular analysis of swine hepatitis E virus from north India.Indian J Med Res. 2010 Nov;132(5):504-8. Indian J Med Res. 2010. PMID: 21149998 Free PMC article.
-
Classification of the Zoonotic Hepatitis E Virus Genotype 3 Into Distinct Subgenotypes.Front Microbiol. 2021 Jan 28;11:634430. doi: 10.3389/fmicb.2020.634430. eCollection 2020. Front Microbiol. 2021. PMID: 33584599 Free PMC article.
-
Hepatitis E virus epidemiology in industrialized countries.Emerg Infect Dis. 2003 Apr;9(4):448-54. doi: 10.3201/eid0904.020351. Emerg Infect Dis. 2003. PMID: 12702225 Free PMC article.
References
-
- Arankall, V. A., L. P. Chobe, M. V. Joshi, M. S. Chadha, B. Kundu, and A. M. Walimbe. 2002. Human and swine hepatitis E viruses from western India belong to different genotypes. J. Hepatol. 36:417-425. - PubMed
-
- Berke, T., and D. O. Mastson. 2000. Reclassification of the Caliciviridae into distinct genera and exclusion of hepatitis A virus from the family on the basis of comparative phylogenetic analysis. Arch. Virol. 145:1421-1436. - PubMed
-
- Chandler, J. D., M. A. Riddell, F. Li, R. J. Love, and D. A. Anderson. 1999. Serological evidence for swine hepatitis E virus infection in Australian pig herds. Vet. Microbiol. 68:95-105. - PubMed
-
- Chaudhary, R. K., M. Morbey, and P. O. Yarbough. 1995. Detection of hepatitis E virus antibody in Canada. Can. J. Infect. Dis. 6:264-265.
-
- Erker, J. C., S. M. Desai, G. G. Schlauder, G. J. Dawson, and I. K. Mushahwar. 1999. A hepatitis E virus variant from the United States: molecular characterization and transmission in Cynomolgus macaques. J. Gen. Virol. 80:681-690. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

