A versatile communication module for controlling RNA folding and catalysis
- PMID: 12409449
- PMCID: PMC135812
- DOI: 10.1093/nar/gkf596
A versatile communication module for controlling RNA folding and catalysis
Abstract
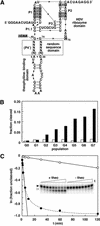
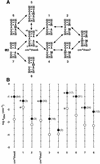
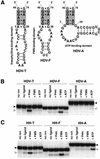
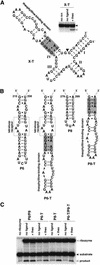
To exert control over RNA folding and catalysis, both molecular engineering strategies and in vitro selection techniques have been applied toward the development of allosteric ribozymes whose activities are regulated by the binding of specific effector molecules or ligands. We now describe the isolation and characterization of a new and considerably versatile RNA element that functions as a communication module to render disparate RNA folding domains interdependent. In contrast to some existing communication modules, the novel 9-nt RNA element is demonstrated to function similarly between a variety of catalysts that include the hepatitis delta virus, hammerhead, X motif and Tetrahymena group I ribozymes, and various ligand-binding domains. The data support a mechanistic model of RNA folding in which the element is comprised of both canonical and non-canonical base pairs and an unpaired nucleotide in the active, effector-bound conformation. Aside from enabling effector-controlled RNA function through rational design, the element can be utilized to identify sites in large RNAs that are susceptible to effector regulation.
Figures
References
-
- Tang J. and Breaker,R.R. (1997) Rational design of allosteric ribozymes. Chem. Biol., 4, 453–459. - PubMed
-
- Kuwabara T., Warashina,M., Tanabe,T., Kenzaburo,T., Asano,S. and Taira,K. (1998) A novel allosterically trans-activated ribozyme, the maxizyme, with exceptional specificity in vitro and in vivo. Mol. Cell, 2, 617–627. - PubMed
-
- Robertson M.P. and Ellington,A.D. (1999) In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol., 17, 62–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

