Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control
- PMID: 12409452
- PMCID: PMC135822
- DOI: 10.1093/nar/gkf602
Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control
Abstract
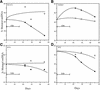
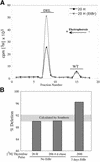
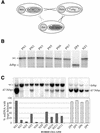
Partially-deleted mitochondrial DNA (DeltamtDNA) accumulates during aging of postmitotic tissues. This accumulation has been linked to decreased metabolic activity, increased reactive oxygen species formation and the aging process. Taking advantage of cell lines with heteroplasmic mtDNA mutations, we showed that, after severe mtDNA depletion, organelles are quickly and predominantly repopulated with DeltamtDNA, whereas repopulation with the wild-type counterpart is slower. This behavior was not observed for full-length genomes with pathogenic point mutations. The faster repopulation of smaller molecules was supported by metabolic labeling of mtDNA with [3H]thymidine during relaxed copy number control conditions. We also showed that hybrid cells containing two defective mtDNA haplotypes tend to retain the smaller one as they adjust their normal mtDNA copy number. Taken together, our results indicate that, under relaxed copy number control, DeltamtDNAs repopulate mitochondria more efficiently than full-length genomes.
Figures



References
-
- Shadel G.S. and Clayton,D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. - PubMed
-
- Moraes C.T. (2001) What regulates mitochondrial DNA copy number in animal cells? Trends Genet., 17, 199–205. - PubMed
-
- Wallace D.C. (1997) Mitochondrial DNA in aging and disease. Sci. Am., 277, 40–47. - PubMed
-
- Raha S. and Robinson,B.H. (2000) Mitochondria, oxygen free radicals, disease and ageing. Trends Biochem. Sci., 25, 502–508. - PubMed
-
- Corral-Debrinski M., Horton,T., Lott,M.T., Shoffner,J.M., Beal,M.F. and Wallace,D.C. (1992) Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nature Genet., 2, 324–329. - PubMed

