Molecular characterization of mouse gastric epithelial progenitor cells
- PMID: 12409607
- PMCID: PMC137502
- DOI: 10.1073/pnas.192574799
Molecular characterization of mouse gastric epithelial progenitor cells
Abstract
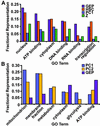
The adult mouse gastric epithelium undergoes continuous renewal in discrete anatomic units. Lineage tracing studies have previously disclosed the morphologic features of gastric epithelial lineage progenitors (GEPs), including those of the presumptive multipotent stem cell. However, their molecular features have not been defined. Here, we present the results of an analysis of genes and pathways expressed in these cells. One hundred forty-seven transcripts enriched in GEPs were identified using an approach that did not require physical disruption of the stem cell niche. Real-time quantitative RT-PCR studies of laser capture microdissected cells retrieved from this niche confirmed enriched expression of a selected set of genes from the GEP list. An algorithm that allows quantitative comparisons of the functional relatedness of automatically annotated expression profiles showed that the GEP profile is similar to a dataset of genes that defines mouse hematopoietic stem cells, and distinct from the profiles of two differentiated GEP descendant lineages (parietal and zymogenic cell). Overall, our analysis revealed that growth factor response pathways are prominent in GEPs, with insulin-like growth factor appearing to play a key role. A substantial fraction of GEP transcripts encode products required for mRNA processing and cytoplasmic localization, including numerous homologs of Drosophila genes (e.g., Y14, staufen, mago nashi) needed for axis formation during oogenesis. mRNA targeting proteins may help these epithelial progenitors establish differential communications with neighboring cells in their niche.
Figures



Comment in
-
Zeroing in on the "holy grail".Gastroenterology. 2003 Jul;125(1):266-7; discussion 267. doi: 10.1016/s0016-5085(03)00831-x. Gastroenterology. 2003. PMID: 12851898 No abstract available.
References
-
- Karam S. M. & Leblond, C. P. (1993) Anat. Rec. 236, 259-279. - PubMed
-
- Karam S. M. & Leblond, C. P. (1993) Anat. Rec. 236, 280-296. - PubMed
-
- Karam S. M. & Leblond, C. P. (1993) Anat. Rec. 236, 297-313. - PubMed
-
- Karam S. M. (1993) Anat. Rec. 236, 314-332. - PubMed
-
- Li Q., Karam, S. M. & Gordon, J. I. (1996) J. Biol. Chem. 271, 3671-3676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

