Plo1(+) regulates gene transcription at the M-G(1) interval during the fission yeast mitotic cell cycle
- PMID: 12411492
- PMCID: PMC131061
- DOI: 10.1093/emboj/cdf564
Plo1(+) regulates gene transcription at the M-G(1) interval during the fission yeast mitotic cell cycle
Abstract
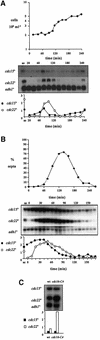
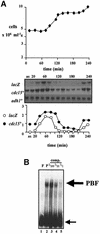
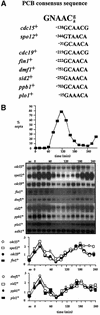
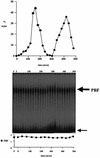
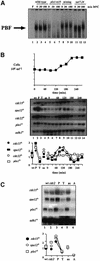
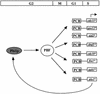
The regulation of gene expression plays an important part in cell cycle controls. We describe the molecular machinery that co-ordinates gene transcription at the M-G(1) interval during the fission yeast mitotic cell cycle. A sequence is identified in the cdc15(+) promoter that we call a PCB (pombe cell cycle box), which confers M-G(1)-specific transcription. Sequences similar to the PCB are present in the promoters of seven other genes, spo12(+), cdc19(+), fin1(+), sid2(+), ppb1(+), mid1(+)/dmf1(+) and plo1(+), which we find to be transcribed at M-G(1). A transcription factor complex is identified that binds to the PCB sequence, which we name PBF, for PCB-binding factor. Finally, we show that PBF binding activity and consequent gene transcription are regulated by the Plo1p protein kinase, thus invoking a potential auto-feedback loop mechanism that regulates mitotic gene transcription and passage through septation and cytokinesis.
Figures



References
-
- Bardin A.J. and Amon,A. (2001) Men and sin: what’s the difference? Nat. Rev. Mol. Cell Biol., 2, 815–826. - PubMed
-
- Broek D., Bartlett,R., Crawford,K. and Nurse,P. (1991) Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature, 349, 388–393. - PubMed
-
- Caligiuri M. and Beach,D. (1993) Sct1 functions in partnership with Cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell, 72, 607–619. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

