Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs
- PMID: 12411503
- PMCID: PMC131082
- DOI: 10.1093/emboj/cdf585
Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs
Abstract
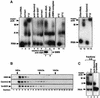
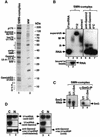
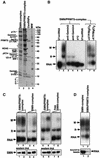
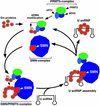
Although spliceosomal Sm proteins can assemble spontaneously onto UsnRNA in vitro, this process requires assisting factors in vivo. SMN, the protein involved in spinal muscular atrophy, is part of a complex that contains the Sm proteins and serves as a critical factor for this reaction. Here, we have reconstituted the SMN-dependent assembly of UsnRNPs in vitro. We demonstrate that the SMN complex is necessary and sufficient for the assembly reaction. The PRMT5 complex, previously implicated in methylation and storage of Sm proteins, interacts with the SMN complex and enhances its activity in an ATP-dependent manner. These data uncover the SMN-PRMT5 complex as a functional entity that promotes the assisted assembly of spliceosomal UsnRNPs, and potentially other, RNA-protein complexes.
Figures


References
-
- Buhler D., Raker,V., Luhrmann,R. and Fischer,U. (1999) Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet., 8, 2351–2357. - PubMed
-
- Campbell L., Hunter,K.M., Mohaghegh,P., Tinsley,J.M., Brasch,M.A. and Davies,K.E. (2000) Direct interaction of Smn with dp103, a putative RNA helicase: a role for Smn in transcription regulation? Hum. Mol. Genet., 9, 1093–1100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

