Ribonuclease activity and RNA binding of recombinant human Dicer
- PMID: 12411504
- PMCID: PMC131075
- DOI: 10.1093/emboj/cdf578
Ribonuclease activity and RNA binding of recombinant human Dicer
Abstract
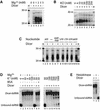
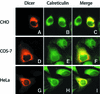
RNA silencing phenomena, known as post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference (RNAi) in animals, are mediated by double-stranded RNA (dsRNA) and mechanistically intersect at the ribonuclease Dicer. Here, we report cloning and expression of the 218 kDa human Dicer, and characterization of its ribonuclease activity and dsRNA-binding properties. The recombinant enzyme generated approximately 21-23 nucleotide products from dsRNA. Processing of the microRNA let-7 precursor by Dicer produced an apparently mature let-7 RNA. Mg(2+) was required for dsRNase activity, but not for dsRNA binding, thereby uncoupling these reaction steps. ATP was dispensable for dsRNase activity in vitro. The Dicer.dsRNA complex formed at high KCl concentrations was catalytically inactive, suggesting that ionic interactions are involved in dsRNA cleavage. The putative dsRNA-binding domain located at the C-terminus of Dicer was demonstrated to bind dsRNA in vitro. Human Dicer expressed in mammalian cells colocalized with calreticulin, a resident protein of the endoplasmic reticulum. Availability of the recombinant Dicer protein will help improve our understanding of RNA silencing and other Dicer-related processes.
Figures





References
-
- Abou Elela S., Igel,H. and Ares,M.,Jr (1996) RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell, 85, 115–124. - PubMed
-
- Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. - PubMed
-
- Baulcombe D.C. (1996) RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol. Biol., 32, 79–88. - PubMed
-
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

