Amino acid residues in the carboxy-terminal region of cottontail rabbit papillomavirus E6 influence spontaneous regression of cutaneous papillomas
- PMID: 12414922
- PMCID: PMC136889
- DOI: 10.1128/jvi.76.23.11801-11808.2002
Amino acid residues in the carboxy-terminal region of cottontail rabbit papillomavirus E6 influence spontaneous regression of cutaneous papillomas
Abstract
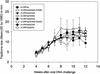
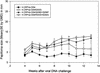
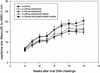
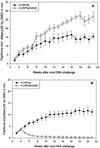
Previous studies have identified two different strains of cottontail rabbit papillomavirus (CRPV) that differ by approximately 5% in base pair sequence and that perform quite differently when used to challenge New Zealand White (NZW) rabbit skin. One strain caused persistent lesions (progressor strain), and the other induced papillomas that spontaneously regressed (regressor strain) at high frequencies (J. Salmon, M. Nonnenmacher, S. Caze, P. Flamant, O. Croissant, G. Orth, and F. Breitburd, J. Virol. 74:10766-10777, 2000; J. Salmon, N. Ramoz, P. Cassonnet, G. Orth, and F. Breitburd, Virology 235:228-234, 1997). We generated a panel of CRPV genomes that contained chimeric and mutant progressor and regressor strain E6 genes and assessed the outcome upon infection of both outbred and EIII/JC inbred NZW rabbits. The carboxy-terminal 77-amino-acid region of the regressor CRPV strain E6, which contained 15 amino acid residues that are different from those of the equivalent region of the persistent CRPV strain E6, played a dominant role in the conversion of the persistent CRPV strain to one showing high rates of spontaneous regressions. In addition, a single amino acid change (G252E) in the E6 protein of the CRPV progressor strain led to high frequencies of spontaneous regressions in inbred rabbits. These observations imply that small changes in the amino acid sequences of papillomavirus proteins can dramatically impact the outcome of natural host immune responses to these viral infections. The data imply that intrastrain differences between separate isolates of a single papillomavirus type (such as human papillomavirus type 16) may contribute to a collective variability in host immune responses in outbred human populations.
Figures


References
-
- Apple, R. J., H. A. Erlich, W. Klitz, M. M. Manos, T. M. Becker, and C. M. Wheeler. 1994. HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat. Genet. 6:157-162. - PubMed
-
- Beekman, N. J., P. A. van Veelen, T. van Hall, A. Neisig, A. Sijts, M. Camps, P. M. Kloetzel, J. J. Neefjes, C. J. Melief, and F. Ossendorp. 2000. Abrogation of CTL epitope processing by single amino acid substitution flanking the C-terminal proteasome cleavage site. J. Immunol. 164:1898-1905. - PubMed
-
- Beskow, A. H., A. M. Josefsson, and U. B. Gyllensten. 2001. HLA class II alleles associated with infection by HPV16 in cervical cancer in situ. Int. J. Cancer 93:817-822. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

