Inhibition of polyomavirus ori-dependent DNA replication by mSin3B
- PMID: 12414923
- PMCID: PMC136908
- DOI: 10.1128/jvi.76.23.11809-11818.2002
Inhibition of polyomavirus ori-dependent DNA replication by mSin3B
Abstract
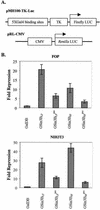
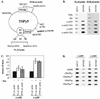
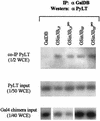
When tethered in cis to DNA, the transcriptional corepressor mSin3B inhibits polyomavirus (Py) ori-dependent DNA replication in vivo. Histone deacetylases (HDACs) appear not to be involved, since tethering class I and class II HDACs in cis does not inhibit replication and treating the cells with trichostatin A does not specifically relieve inhibition by mSin3B. However, the mSin3B L59P mutation that impairs mSin3B interaction with N-CoR/SMRT abrogates inhibition of replication, suggesting the involvement of N-CoR/SMRT. Py large T antigen interacts with mSin3B, suggesting an HDAC-independent mechanism by which mSin3B inhibits DNA replication.
Figures





References
-
- Alexiadis, V., L. Halmer, and C. Gruss. 1997. Influence of core histone acetylation on SV40 minichromosome replication in vitro. Chromosoma 105:324-331. - PubMed
-
- Alland, L., R. Muhle, H. Hou, J. Potes, L. Chin, N. Schreiber-Agus, and R. A. Depinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. - PubMed
-
- Amin, A. A., Y. Murakami, and J. Hurwitz. 1994. Initiation of DNA replication by simian virus 40 T antigen is inhibited by the p107 protein. J. Biol. Chem. 269:7735-7743. - PubMed
-
- Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with Siners and NuRDs. Trends Cell Biol. 9:193-198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

