Cell-free replication of the hepatitis C virus subgenomic replicon
- PMID: 12414942
- PMCID: PMC136887
- DOI: 10.1128/jvi.76.23.12001-12007.2002
Cell-free replication of the hepatitis C virus subgenomic replicon
Abstract
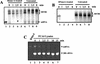
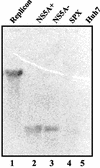
The hepatitis C virus (HCV) contains a plus-strand RNA genome. The 5' noncoding region (NCR) of the viral genome functions as an internal ribosome entry site, and its unique 3' NCR is required for the assembly of the replication complex during initiation of HCV RNA replication. Lohmann et al. (V. Lohmann, F. Korner, J.-O. Koch, U. Herian, L. Theilman, and R. Batenschlager, Science 285:110-113, 1999) developed a subgenomic HCV replicon system, which represents an important tool in studying HCV replication in cultured cells. In this study, we describe a cell-free replication system that utilizes cytoplasmic lysates prepared from Huh-7 cells harboring the HCV subgenomic replicons. These lysates, which contain ribonucleoprotein complexes associated with cellular membranes, were capable of incorporating [alpha(32)P]CTP into newly synthesized RNA from subgenomic replicons in vitro. Replicative forms (RFs) and replicative intermediates (RIs) were synthesized from the endogenous HCV RNA templates. Consistent with previous observations, RFs were found to be resistant to RNase A digestion, whereas RIs were sensitive to RNase treatment. The radiolabeled HCV RF-RI complexes contained both minus and plus strands and were specific to the lysates derived from replicon-expressing cells. The availability of a cell-free replication system offers opportunities to probe the mechanism(s) of HCV replication. It also provides a novel assay for potential therapeutic agents.
Figures



References
-
- Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, M. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold Des. 7:1417-1426. - PubMed
-
- Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. - PubMed
-
- Bartholomeusz, A. I., and P. J. Wright. 1993. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch. Virol. 128:111-121. - PubMed
-
- Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

