Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses
- PMID: 12414947
- PMCID: PMC136909
- DOI: 10.1128/jvi.76.23.12055-12068.2002
Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses
Abstract
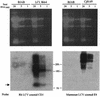
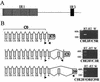
Callitrichine herpesvirus 3 (CalHV-3) was isolated from a B-cell lymphoma arising spontaneously in the New World primate Callithrix jacchus, the common marmoset. Partial genomic sequence analysis definitively identified CalHV-3 as a member of the Epstein-Barr virus (EBV)-related lymphocryptovirus (LCV) genus and extended the known host range of LCVs beyond humans and Old World nonhuman primates. We have now completed the first genomic sequence of an LCV infecting a New World primate by describing the unique short region, the major internal repeat, and a portion of the unique long region. This portion of the genome contains the putative latent origin of replication and 13 additional open reading frames (ORFs), 5 of which show no homology to any viral or cell genes. One of the novel genes, C5, is a positional homologue for the transformation-essential EBV gene EBNA-2. The marmoset LCV genome is also notable for the absence of viral interleukin-10 and small nonpolyadenylated RNA homologues. Marmoset LCV transcripts encoding putative latent infection nuclear proteins have a common leader sequence that is spliced from the major internal repeat in a manner similar to that of the EBV EBNA-LP, suggesting strong conservation of a common promoter and splicing of these latent infection mRNAs. An EBV LMP2A-like spliced transcript crossing the terminal repeats encodes a unique ORF, C7, with multiple transmembrane domains and tyrosine kinase phosphorylation sites functionally reminiscent of EBV LMP2A. However, the carboxy-terminal location of the candidate phosphotyrosine residues is more reminiscent of the Kaposi's sarcoma-associated herpesvirus K15 gene and provides potential evidence of an evolutionary transition from rhadinoviruses to lymphocryptoviruses. The unusual gene repertoire of the marmoset LCV differentiates ancestral viral genes likely present in an LCV progenitor from viral genes acquired later as primates and LCV coevolved, providing a defining point in the evolution of oncogenic LCVs.
Figures





References
-
- Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

