Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import
- PMID: 12414950
- PMCID: PMC136890
- DOI: 10.1128/jvi.76.23.12087-12096.2002
Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import
Abstract
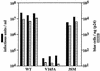
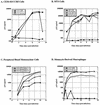
Human immunodeficiency virus type 1 (HIV-1) can infect nondividing cells productively because the nuclear import of viral nucleic acids occurs in the absence of cell division. A number of viral factors that are present in HIV-1 preintegration complexes (PICs) have been assigned functions in nuclear import, including an essential valine at position 165 in integrase (IN-V165) and the central polypurine tract (cPPT). In this article, we report a comparison of the replication and infection characteristics of viruses with disruptions in the cPPT and IN-V165. We found that viruses with cPPT mutations still replicated productively in both dividing and nondividing cells, while viruses with a mutation at IN-V165 did not. Direct observation of the subcellular localization of HIV-1 cDNAs by fluorescence in situ hybridization revealed that cDNAs synthesized by both mutant viruses were readily detected in the nucleus. Thus, neither the cPPT nor the valine residue at position 165 of integrase is essential for the nuclear import of HIV-1 PICs.
Figures




References
-
- Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. - PubMed
-
- Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347-356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

